Another fantastic year of discovery is over – read on for our 2016 plant science top picks!
January

A Zostera marina meadow in the Archipelago Sea, southwest Finland. Image credit: Christoffer Boström (Olsen et al., 2016. Nature).
The year began with the publication of the fascinating eelgrass (Zostera marina) genome by an international team of researchers. This marine monocot descended from land-dwelling ancestors, but went through a dramatic adaptation to life in the ocean, in what the lead author Professor Jeanine Olsen described as, “arguably the most extreme adaptation a terrestrial… species can undergo”.
One of the most interesting revelations was that eelgrass cannot make stomatal pores because it has completely lost the genes responsible for regulating their development. It also ditched genes involved in perceiving UV light, which does not penetrate well through its deep water habitat.
Read the paper in Nature: The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea.
BLOG: You can find out more about the secrets of seagrass in our blog post.
February
Plants are known to form new organs throughout their lifecycle, but it was not previously clear how they organized their cell development to form the right shapes. In February, researchers in Germany used an exciting new type of high-resolution fluorescence microscope to observe every individual cell in a developing lateral root, following the complex arrangement of their cell division over time.
Using this new four-dimensional cell lineage map of lateral root development in combination with computer modelling, the team revealed that, while the contribution of each cell is not pre-determined, the cells self-organize to regulate the overall development of the root in a predictable manner.
Watch the mesmerizing cell division in lateral root development in the video below, which accompanied the paper:
Read the paper in Current Biology: Rules and self-organizing properties of post-embryonic plant organ cell division patterns.
March
In March, a Spanish team of researchers revealed how the anti-wilting molecular machinery involved in preserving cell turgor assembles in response to drought. They found that a family of small proteins, the CARs, act in clusters to guide proteins to the cell membrane, in what author Dr. Pedro Luis Rodriguez described as “a kind of landing strip, acting as molecular antennas that call out to other proteins as and when necessary to orchestrate the required cellular response”.
Read the paper in PNAS: Calcium-dependent oligomerization of CAR proteins at cell membrane modulates ABA signaling.
*If you’d like to read more about stress resilience in plants, check out the meeting report from the Stress Resilience Forum run by the GPC in coalition with the Society for Experimental Biology.*
April
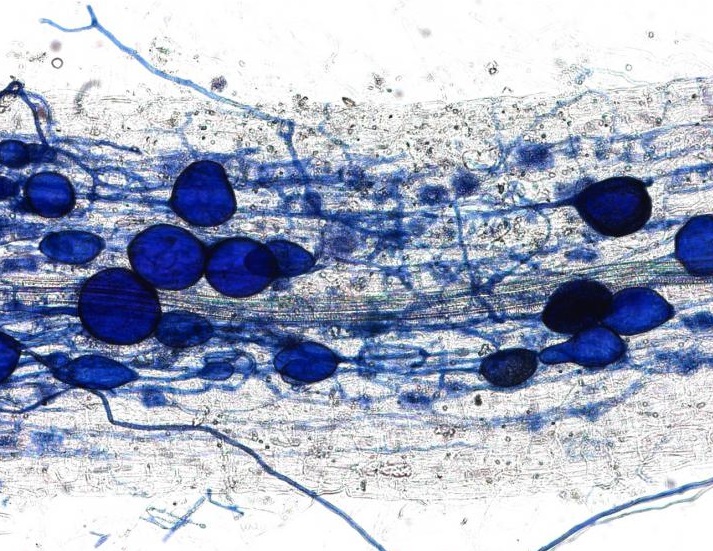
This plant root is infected with arbuscular mycorrhizal fungi. Image credit: University of Zurich.
In April, we received an amazing insight into the ‘decision-making ability’ of plants when a Swiss team discovered that plants can punish mutualist fungi that try to cheat them. In a clever experiment, the researchers provided a plant with two mutualistic partners; a ‘generous’ fungus that provides the plant with a lot of phosphates in return for carbohydrates, and a ‘meaner’ fungus that attempts to reduce the amount of phosphate it ‘pays’. They revealed that the plants can starve the meaner fungus, providing fewer carbohydrates until it pays its phosphate bill.
Author Professor Andres Wiemsken explains: “The plant exploits the competitive situation of the two fungi in a targeted manner, triggering what is essentially a market-based process determined by cost and performance”.
Read the paper in Ecology Letters: Options of partners improve carbon for phosphorus trade in the arbuscular mycorrhizal mutualism.
May
The transition of ancient plants from water onto land was one of the most important events in our planet’s evolution, but required a massive change in plant biology. Suddenly plants risked drying out, so had to develop new ways to survive drought.
In May, an international team discovered a key gene in moss (Physcomitrella patens) that allows it to tolerate dehydration. This gene, ANR, was an ancient adaptation of an algal gene that allowed the early plants to respond to the drought-signaling hormone ABA. Its evolution is still a mystery, though, as author Dr. Sean Stevenson explains: “What’s interesting is that aquatic algae can’t respond to ABA: the next challenge is to discover how this hormone signaling process arose.”
Read the paper in The Plant Cell: Genetic analysis of Physcomitrella patens identifies ABSCISIC ACID NON-RESPONSIVE, a regulator of ABA responses unique to basal land plants and required for desiccation tolerance.
June
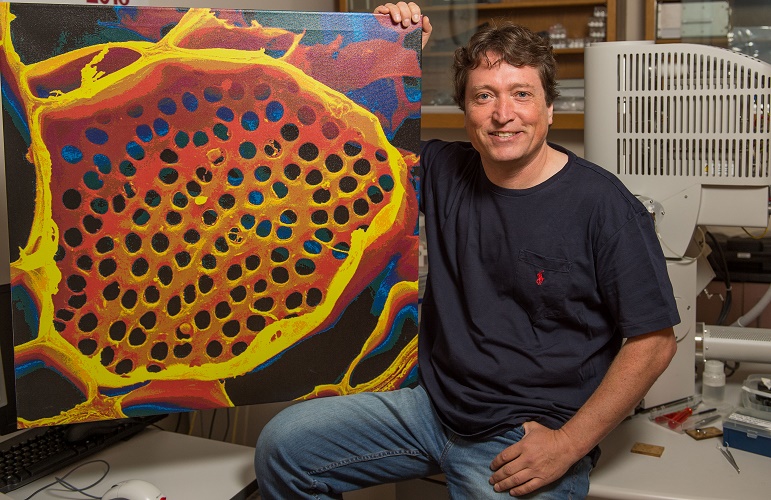
Professor Michael Knoblauch shows off a microscope image of phloem tubes. Image credit: Washington State University.
Sometimes revisiting old ideas can pay off, as a US team revealed in June. In 1930, Ernst Münch hypothesized that transport through the phloem sieve tubes in the plant vascular tissue is driven by pressure gradients, but no-one really knew how this would account for the massive pressure required to move nutrients through something as large as a tree.
Professor Michael Knoblauch and colleagues spent decades devising new methods to investigate pressures and flow within phloem without disrupting the system. He eventually developed a suite of techniques, including a picogauge with the help of his son, Jan, to measure tiny pressure differences in the plants. They found that plants can alter the shape of their phloem vessels to change the pressure within them, allowing them to transport sugars over varying distances, which provided strong support for Münch flow.
Read the paper in eLife: Testing the Münch hypothesis of long distance phloem transport in plants.
BLOG: We featured similar work (including an amazing video of the wound response in sieve tubes) by Knoblauch’s collaborator, Dr. Winfried Peters, on the blog – read it here!
July

Preserved remains of rope, seeds, reeds and pellets (left), and a desiccated barley grain (right) found at Yoram Cave in the Judean Desert. Credit: Uri Davidovich and Ehud Weiss.
In July, an international and highly multidisciplinary team published the genome of 6,000-year-old barley grains excavated from a cave in Israel, the oldest plant genome reconstructed to date. The grains were visually and genetically very similar to modern barley, showing that this crop was domesticated very early on in our agricultural history. With more analysis ongoing, author Dr. Verena Schünemann predicts that “DNA-analysis of archaeological remains of prehistoric plants will provide us with novel insights into the origin, domestication and spread of crop plants”.
Read the paper in Nature Genetics: Genomic analysis of 6,000-year-old cultivated grain illuminates the domestication history of barley.
BLOG: We interviewed Dr. Nils Stein about this fascinating work on the blog – click here to read more!
August
Another exciting cereal paper was published in August, when an Australian team revealed that C4 photosynthesis occurs in wheat seeds. Like many important crops, wheat leaves perform C3 photosynthesis, which is a less efficient process, so many researchers are attempting to engineer the complex C4 photosynthesis pathway into C3 crops.
This discovery was completely unexpected, as throughout its evolution wheat has been a C3 plant. Author Professor Robert Henry suggested: “One theory is that as [atmospheric] carbon dioxide began to decline, [wheat’s] seeds evolved a C4 pathway to capture more sunlight to convert to energy.”
Read the paper in Scientific Reports: New evidence for grain specific C4 photosynthesis in wheat.
September

Professor Stefan Jansson cooks up “Tagliatelle with CRISPRy fried vegetables”. Image credit: Stefan Jansson.
September marked an historic event. Professor Stefan Jansson cooked up the world’s first CRISPR meal, tagliatelle with CRISPRy fried vegetables (genome-edited cabbage). Jansson has paved the way for CRISPR in Europe; while the EU is yet to make a decision about how CRISPR-edited plants will be regulated, Jansson successfully convinced the Swedish Board of Agriculture to rule that plants edited in a manner that could have been achieved by traditional breeding (i.e. the deletion or minor mutation of a gene, but not the insertion of a gene from another species) cannot be treated as a GMO.
Read more in the Umeå University press release: Umeå researcher served a world first (?) CRISPR meal.
BLOG: We interviewed Professor Stefan Jansson about his prominent role in the CRISPR/GM debate earlier in 2016 – check out his post here.
*You may also be interested in the upcoming meeting, ‘New Breeding Technologies in the Plant Sciences’, which will be held at the University of Gothenburg, Sweden, on 7-8 July 2017. The workshop has been organized by Professor Jansson, along with the GPC’s Executive Director Ruth Bastow and Professor Barry Pogson (Australian National University/GPC Chair). For more info, click here.*
October
Phytochromes help plants detect day length by sensing differences in red and far-red light, but a UK-Germany research collaboration revealed that these receptors switch roles at night to become thermometers, helping plants to respond to seasonal changes in temperature.
Dr Philip Wigge explains: “Just as mercury rises in a thermometer, the rate at which phytochromes revert to their inactive state during the night is a direct measure of temperature. The lower the temperature, the slower phytochromes revert to inactivity, so the molecules spend more time in their active, growth-suppressing state. This is why plants are slower to grow in winter”.
Read the paper in Science: Phytochromes function as thermosensors in Arabidopsis.
November

A fossil ginkgo (Ginkgo biloba) leaf with its modern counterpart. Image credit: Gigascience.
In November, a Chinese team published the genome of Ginkgo biloba¸ the oldest extant tree species. Its large (10.6 Gb) genome has previously impeded our understanding of this living fossil, but researchers will now be able to investigate its ~42,000 genes to understand its interesting characteristics, such as resistance to stress and dioecious reproduction, and how it remained almost unchanged in the 270 million years it has existed.
Author Professor Yunpeng Zhao said, “Such a genome fills a major phylogenetic gap of land plants, and provides key genetic resources to address evolutionary questions [such as the] phylogenetic relationships of gymnosperm lineages, [and the] evolution of genome and genes in land plants”.
Read the paper in GigaScience: Draft genome of the living fossil Ginkgo biloba.
December
The year ended with another fascinating discovery from a Danish team, who used fluorescent tags and microscopy to confirm the existence of metabolons, clusters of metabolic enzymes that have never been detected in cells before. These metabolons can assemble rapidly in response to a stimulus, working as a metabolic production line to efficiently produce the required compounds. Scientists have been looking for metabolons for 40 years, and this discovery could be crucial for improving our ability to harness the production power of plants.
Read the paper in Science: Characterization of a dynamic metabolon producing the defense compound dhurrin in sorghum.
Another amazing year of science! We’re looking forward to seeing what 2017 will bring!
P.S. Check out 2015 Plant Science Round Up to see last year’s top picks!







