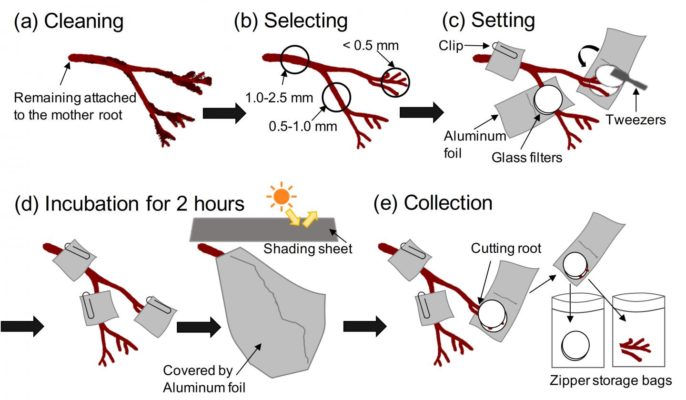
Plants produce the hormone jasmonic acid as a defence response when challenged. This is how they ensure that their predators no longer like the taste of their leaves. Biologists want to find out whether biological precursors and other variants of…
Read More