



One of the world’s most widely used glyphosate-based herbicides, Roundup, can trigger loss of biodiversity, making ecosystems more vulnerable to pollution and climate change, say researchers from McGill University.
The widespread use of Roundup on farms has sparked concerns over potential health and environmental effects globally. Since the 1990s use of the herbicide boomed, as the farming industry adopted “Roundup Ready” genetically modified crop seeds that are resistant to the herbicide. “Farmers spray their corn and soy fields to eliminate weeds and boost production, but this has led to glyphosate leaching into the surrounding environment. In Quebec, for example, traces of glyphosate have been found in Montérégie rivers,” says Andrew Gonzalez, a McGill biology professor and Liber Ero Chair in Conservation Biology.
To test how freshwater ecosystems respond to environmental contamination by glyphosate, researchers used experimental ponds to expose phytoplankton communities (algae) to the herbicide. “These tiny species at the bottom of the food chain play an important role in the balance of a lake’s ecosystem and are a key source of food for microscopic animals. Our experiments allow us to observe, in real time, how algae can acquire resistance to glyphosate in freshwater ecosystems,” says post-doctoral researcher Vincent Fugère.
The researchers found that freshwater ecosystems that experience moderate contamination from the herbicide became more resistant when later exposed to a very high level of it – working as a form of “evolutionary vaccination.” According to the researchers, the results are consistent with what scientists call “evolutionary rescue,” which until recently had only been tested in the laboratory. Previous experiments by the Gonzalez group had shown that evolutionary rescue can prevent the extinction of an entire population when exposed to severe environmental contamination by a pesticide thanks to the rapid evolution.
However, the researchers note that the resistance to the herbicide came at a cost of plankton diversity. “We observed significant loss of biodiversity in communities contaminated with glyphosate. This could have a profound impact on the proper functioning of ecosystems and lower the chance that they can adapt to new pollutants or stressors. This is particularly concerning as many ecosystems are grappling with the increasing threat of pollution and climate change,” says Gonzalez.
The researchers point out that it is still unclear how rapid evolution contributes to herbicide resistance in these aquatic ecosystems. Scientist already know that some plants have acquired genetic resistance to glyphosate in crop fields that are sprayed heavily with the herbicide. Finding out more will require genetic analyses that are currently under way by the team.
Read the paper: Nature Ecology & Evolution
Article source: McGill University
Image credit: Vincent Fugère
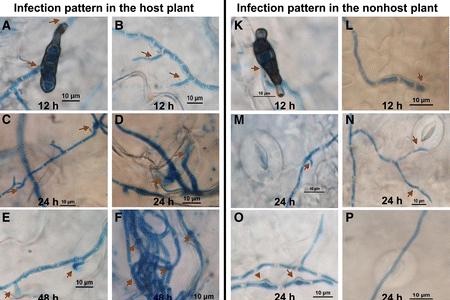

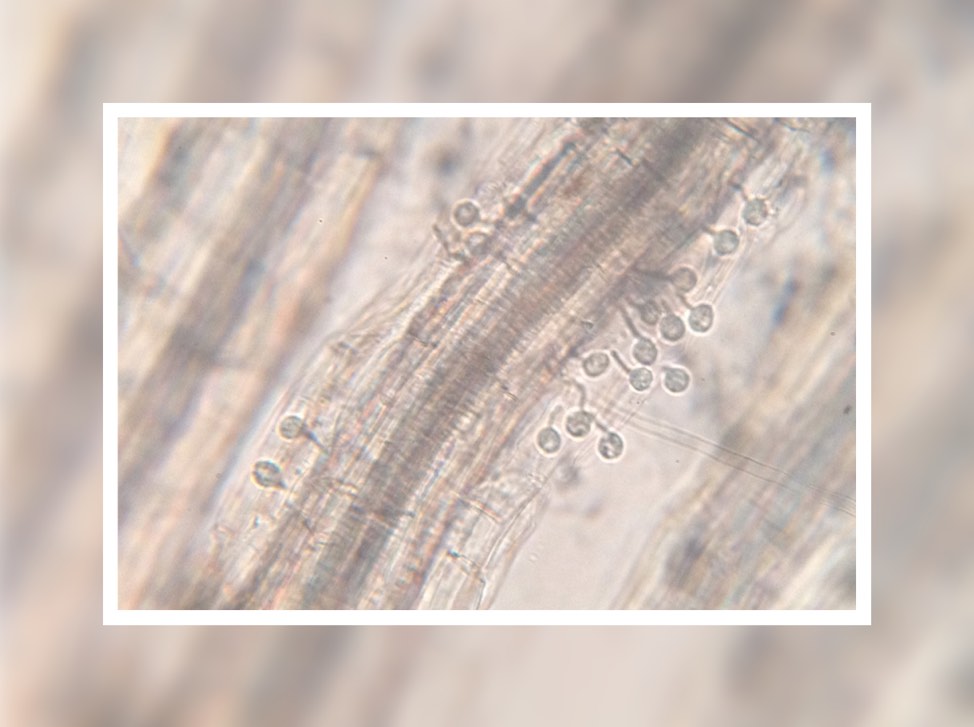

Many New York tomato growers are familiar with the scourge of bacterial canker – the wilted leaves and blistered fruit that can spoil an entire season’s planting. For those whose livelihoods depend on tomatoes, this pathogen – Clavibacter michiganensis – is economically devastating.
In a new paper, Cornell researchers showed that wild tomato varieties are less affected by bacterial canker than traditionally cultivated varieties. The paper, “Characterizing Colonization Patterns of Clavibacter michiganensis During Infection of Tolerant Wild Solanum Species,” published online in the journal Phytopathology.
Co-authors were Christine Smart, professor of plant pathology and plant-microbe biology in the College of Agriculture and Life Sciences; F. Christopher Peritore-Galve, a doctoral student in the Smart Lab; and Christine Miller, a 2018 Smart Lab undergraduate summer intern from North Carolina State University.
“Bacterial canker is pretty bad in New York,” Peritore-Galve said, “but it’s distributed worldwide, everywhere tomatoes are grown.”
The pathogen causes wounding and is spread by wind-blown rain; if one tomato gets infected, it can spread from plant to plant.
“Bacterial canker certainly can cause the complete loss of a field of tomatoes, and we see outbreaks of the disease every year,” Smart said. “Growers use disease management strategies, including spraying plants with copper-based products; however, once there is an outbreak it’s difficult to control bacterial canker.”
To combat diseases, plant pathologists and breeders often look for varieties that are resistant, but among tomatoes traditionally grown for market, there are none with genetic resistance to bacterial canker. So Peritore-Galve, Miller and Smart went back to the beginning.
Tomatoes are native to the Andes Mountains region of South America, where wild species have been free to evolve for thousands of years. Recently, plant breeders have identified wild tomatoes that seem to be less susceptible to bacterial canker and are resistant to other pathogens.
The team wanted to understand how bacteria spread and colonize in wild tomatoes versus cultivated ones. They zeroed in on the plants’ vascular systems – specifically their xylem vessels.
Like individual veins in a human, xylem vessels transport water and nutrients from soil throughout the plant. The team found that in cultivated species, bacterial canker spreads everywhere, while in wild species the bacteria remain confined to certain xylem vessels without moving much into surrounding tissues.
“The wild tomatoes, for some reason, impede the ability of the bacteria to move up and down through the plants, which reduces symptoms – in this case, leaf wilt,” Peritore-Galve said.
This is the first study ever confirming that wild tomatoes are susceptible to bacterial canker, though the infection is less severe than in cultivated varieties. But while a severe infection causes fewer symptoms in the wild plant, it can still cause lesions on the fruit.
Even so, a tomato variety with resistance to the bacteria could still be very helpful for tomato growers, said Chuck Bornt, vegetable specialist with Cornell Cooperative Extension’s Eastern New York Commercial Horticulture program. Bornt works extensively with New York tomato growers.
“Many times, it’s not the fruit symptoms that cause the issue,” Bornt said, “it’s the wilting of the plants or the plugging of the xylem cells that cause the plant to lose foliage, which then exposes the fruit to sun scald and other issues. … Infected fruit are also an issue, but in my opinion it’s these other issues that have more impact.”
Read the paper: Phytopathology
Article source: Cornell University
Author: Krisy Gashler
Image credit: Allison Usavage/Cornell University


While studies of the microbiomes (which comprises all the microorganisms, mainly bacteria and fungi) of the phyllosphere and the rhizosphere of plants are important, scientists at INRA believe more attention should be given to the microbiomes of crop residues.
Crop residues are important as a key microbial ecosystem with the power to contribute both negatively and positively to crop health and productivity. Crop residues are a breeding ground for plant disease but also contribute significantly to the stability of agrosystems.
“Residues deserve special attention in the context of crop protection,” plant pathologist Frédéric Suffert explains. “First, because residues are ‘the problem’ as the main support of pathogens that cause disease. Second because residues can be also part of the solution to control these diseases.”
Focusing on cereal crops, the INRA scientists explored how dynamic interactions between microbial communities of residues can contribute to innovative disease management strategies such as next-generation microbiome-based biocontrol.
“We connected residue microbiome with the survival of residue-borne fungal plant pathogens by combining knowledge in microbial ecology and epidemiology,” explained Frédéric Suffert. “This is the first time this connection has been made.”
Read the paper: Phytobiomes
Article source: American Phytopathologycal Society via Eurekalert
Image credit: Shutterbug75 / Pixabay
