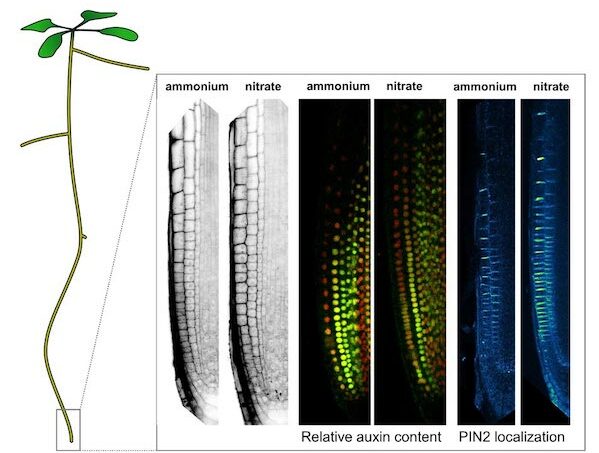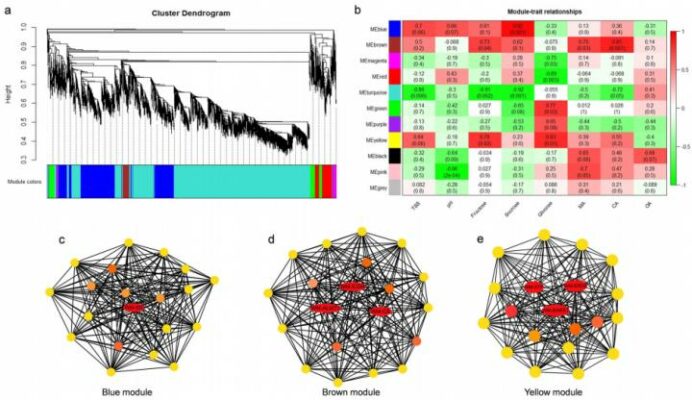
The sensory quality of watermelon fruit is determined by the content of sugar and organic acid, which determines the taste of watermelon during the development and maturation of watermelon fruit. The changes of sugar and organic acid during the watermelon fruit development were analyzed and the key gene networks controlling the metabolism of sugar and organic acid during the fruit development were identified.