Most of the world’s food is produced in temperate zones. The Global Food Security program’s Evangelia Kougioumoutzi reports on the TempAg network.
Agricultural production in temperate regions is highly productive with a significant proportion of global output originating from temperate (i.e. non-tropical) countries – 21% of global meat production and 20% of global cereal production [link opens PDF] originate from Europe alone. This proportion is very likely to increase in light of climate change.

Little fluffy clouds: temperate zones are well suited to agricultural production. Image credit: connect11/Thinkstock
TempAg is an international research collaboration network that was established to increase the impact of agricultural research and inform policy making in the world’s temperate regions. Its work does not solely focus on research, but also provides insights into current thinking through mapping existing scientific findings and outstanding knowledge gaps. In this way, the network aspires to become a platform for the alignment of national agricultural research and food partnership programs (such as Global Food Security) that will enable the development of more effective agricultural policies with a long-term vision.
Since its official inauguration in Paris in April 2015, TempAg has been leading a series of on-going workstreams around:
- Boosting resilience of agricultural production systems at multiple scales and levels
- Optimising land management for ecosystem services and food production
- Improving sustainability of food productivity in the farms & enterprise level
You can read more about these themes on the TempAg website: http://tempag.net/themes/.
Future foresights
After 18 months of existence, TempAg held a foresight workshop in London on 5–7 October to determine its future priorities.
Forty delegates took part in the workshop, coming from the 14 different countries in the temperate region, and from academia, policy, industry, and professionals at the science–policy interface. Through a series of presentations and interactive sessions, participants were invited to consider what the current and future challenges are in temperate agriculture, taking into account the needs of policy makers and industry in helping them to improve sustainable agriculture practices.
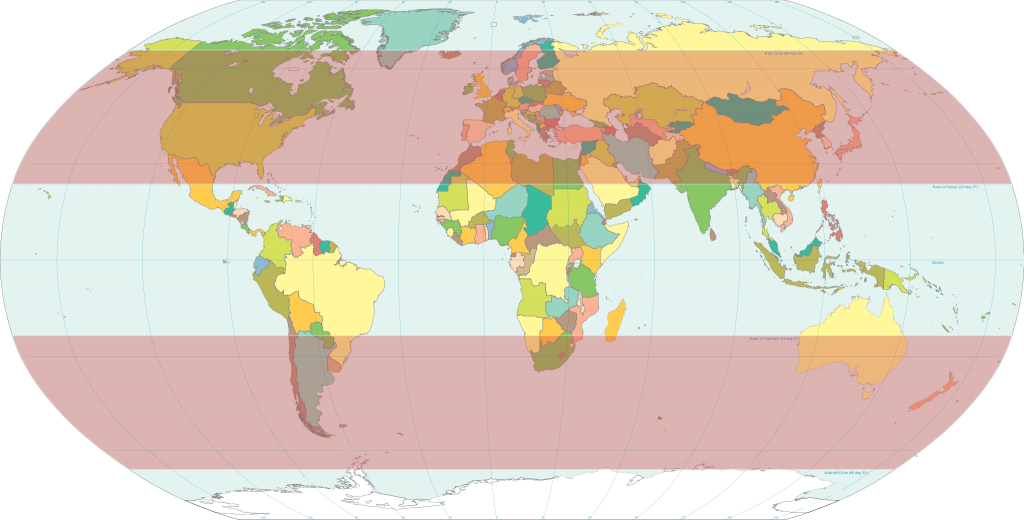
Temperate zones cover much of the world’s major food-growing areas. Image from Wikipedia/CIA-Factbook
To tackle sustainability in temperate agriculture, there is a need to better manage risks and stresses (both biotic and abiotic), as well as finding ways to manage the restoration of natural capital, ecosystem services, and soils. During the workshop, it was noted that utilizing the diversity within different agricultural systems, via identifying the best practice and using the appropriate technological mix, may be a way forward in making production systems more sustainable.
Participants stressed the importance of taking a holistic view of the sustainability agenda within agriculture, without just focusing on environmental aspects. This means also taking into consideration socioeconomic factors, such as making food value chains (like turning milk into cheese), more equitable by identifying who gets the equity from the food commodities’ prices, or identifying what the optimum legal framework for sharing data might be.
The group also considered sustainable agriculture issues from a policy and industry needs angle. It was interesting to see that dealing with shocks (environmental, socioeconomic, and technological) featured highly in this discussion as well. It was suggested that increasing resilience to these shocks could be facilitated via the widespread diffusion of existing technologies. Engaging with farmers during this time would be necessary to identify technology uptake barriers.
Forward moves
Future-proofing agricultural resilience and enhancing the capacity to respond to shocks was deemed an urgent priority, so the development of a comprehensive map identifying the multiple shocks that could impact on farm resilience in temperate zones might be a future workstream for TempAg. Work in this area could help develop models to assess the flexibility within agricultural production systems.

Click to visit the TempAg website. Image: TempAg
What we eat is largely based on the types of food we produce. Therefore, healthy diets are intrinsically linked with our production systems. Another area of interest for TempAg could be to explore what the nutritional value of crops should be for better health, and what a nutritional diet will look like for sustainable temperate agriculture. Developing frameworks in this area could further inform future farming practices in temperate areas.
Since TempAg’s initiation, two major global policy agendas have been adopted by the international community: the Sustainable Development Goals and the Paris COP21 agreement. Identifying what types of data and scientific evidence policy makers will need to achieve the agriculture-relevant targets was another area where TempAg could focus its activity moving forward.
Finally, delegates highlighted areas of work that could help to build more effective policies with a longer-term vision. These included developing economic tools for valuing natural capital and ecosystem services, as well as integrated assessment tools to monitor the performance and impact (environmental cost) of existing policies.
This article is cross-posted with the Global Food Security blog.
About Evangelia Kougioumoutzi
Evangelia is International Coordinator & Programme Manager for the Global Food Security program (GFS). Before joining GFS, Evangelia worked as an Innovation Manager for GFS partners BBSRC. She holds a PhD in plant development and genetics from the University of Oxford.