This week’s blog post was written by Dr Damiano Martignago, a genome editing specialist at Rothamsted Research.
Genome editing technologies comprise a diverse set of molecular tools that allow the targeted modification of a DNA sequence within a genome. Unlike “traditional” breeding, genome editing does not rely on random DNA recombination; instead it allows the precise targeting of specific DNA sequences of interest. Genome editing approaches induce a double strand break (DSB) of the DNA molecule at specific sites, activating the cell’s DNA repair system. This process could be either error-prone, thus used by scientists to deactivate “undesired” genes, or error-free, enabling target DNA sequences to be “re-written” or the insertion of DNA fragments in a specific genomic position.
The most promising among the genome editing technologies, CRISPR/Cas9, was chosen as Science’s 2015 Breakthrough of the Year. Cas9 is an enzyme able to target a specific position of a genome thanks to a small RNA molecule called guide RNA (gRNA). gRNAs are easy to design and can be delivered to cells along with the gene encoding Cas9, or as a pre-assembled Cas9-gRNA protein-RNA complex. Once inside the cell, Cas9 cuts the target DNA sequence homologous to the gRNAs, producing DSBs.
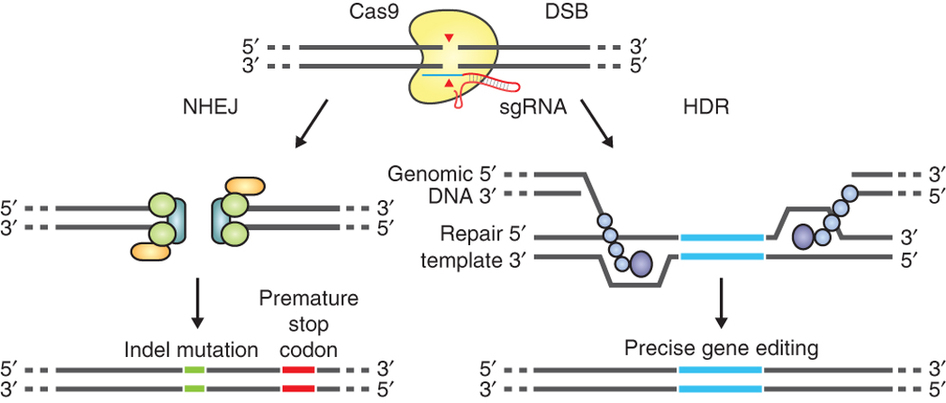
The guide RNA (sgRNA) directs Cas9 to a specific region of the genome, where it induces a double-strand break in the DNA. On the left, the break is repaired by non-homologous-end joining, which can result in insertion/deletion (indel) mutations. On the right, the homologous-directed recombination pathway creates precise changes using a supplied template DNA. Credit: Ran et al. (2013). Nature Protocols.
Genome editing in crops
Together with the increased data availability on crop genomes, genome editing techniques such as CRISPR are allowing scientists to carry out ambitious research on crop plants directly, building on the knowledge obtained during decades of investigation in model plants.
The concept of CRISPR was first tested in crops by generating cultivars that are resistant to herbicides, as this is an easy trait to screen for and identify. One of the first genome-edited crops, a herbicide-resistant oilseed rape produced by Cibus, has already been grown and harvested in the USA in 2015.

Researchers used CRISPR to engineer a wheat variety resistant to powdery mildew (shown here), a major disease of this crop. Image credit: NY State IPM Program. Used under license: CC BY 2.0.
Using CRISPR, scientists from the Chinese Academy of Sciences produced a wheat variety resistant to powdery mildew, one of the major diseases in wheat. Similarly, another Chinese research group exploited CRISPR to produce a rice line with enhanced rice blast resistance that will help to reduce the amount of fungicides used in rice farming. CRISPR/Cas9 has also been already applied to maize, tomato, potato, orange, lettuce, soybean and other legumes.
Genome editing could also revolutionize the management of viral plant disease. The CRISPR/Cas9 system was originally discovered in bacteria, where it provided them with molecular immunity against viruses, but it can also be moved into plants. Scientists can transform plants to produce the Cas9 and gRNAs that target viral DNA, reducing virus accumulation; alternatively, they can suppress those plant genes that are hijacked by the virus to mediate its own diffusion in the plants. Since most plants are defenseless against viruses and there are no chemical controls available for plant viruses, the main method to stop the spread of these diseases is still the destruction of the infected plant. For the first time in history, scientists have an effective weapon to fight back against plant viruses.

The cassava brown streak disease virus can destroy cassava crops, threatening the food security of the 300 million people who rely on this crop in Africa. Image credit: Katie Tomlinson (for more on this topic, read her blog here).
Genome editing will be particularly useful in the genetic improvement of many crops that are propagated mainly by vegetative reproduction, and so very difficult to improve by traditional breeding methods involving crossing (e.g. cassava, banana, grape, potato). For example, using TALENs, scientists from Cellectis edited a potato line to minimize the accumulation of reducing sugars that may be converted into acrylamide (a possible carcinogen) during cooking.
Concerns about off-targets
One of the hypothesized risks of using CRISPR/Cas9 is the potential targeting of undesired DNA regions, called off-targets. It is possible to limit the potential for off-targets by designing very specific gRNAs, and all of the work published so far either did not detect any off-targets or, if detected, they occurred at a very low frequency. The number of off-target mutations produced by CRISPR/Cas9 is therefore minimal, especially if compared with the widely accepted random mutagenesis of crops used in plant breeding since the 1950s.
GM or not-GM
Genome editing is interesting from a regulatory point of view too. After obtaining the desired heritable mutation using CRISPR/Cas9, it is possible to remove the CRISPR/Cas9 integrated vectors from the genome using simple genetic segregation, leaving no trace of the genome modification other than the mutation itself. This means that some countries (including the USA, Canada, and Argentina) consider the products of genome editing on a case-by-case basis, ruling that a crop is non-GM when it contains gene combinations that could have been obtained through crossing or random mutation. Many other countries are yet to issue an official statement on CRISPR, however.
Recently, scientists showed that is possible to edit the genome of plants without adding any foreign DNA and without the need for bacteria- or virus-mediated plant transformation. Instead, a pre-assembled Cas9-gRNA ribonucleoprotein (RNP) is delivered to plant cells in vitro, which can edit the desired region of the genome before being rapidly degraded by the plant endogenous proteases and nucleases. This non-GM approach can also reduce the potential of off-target editing, because of the minimal time that the RNP is present inside the cell before being degraded. RNP-based genome editing has been already applied to tobacco plants, rice, and lettuce, as well as very recently to maize.
In conclusion, genome editing techniques, and CRISPR/Cas9 in particular, offers scientists and plant breeders a flexible and relatively easy approach to accelerate breeding practices in a wide variety of crop species, providing another tool that we can use to improve food security in the future.
For more on CRISPR, check out this recent TED Talk from Ellen Jorgensen:
About the author
Dr Damiano Martignago is a plant molecular biologist who graduated from Padua University, Italy, with a degree in Food Biotechnology in 2009. He obtained his PhD in Biology at Roma Tre University in 2014. His experience with CRISPR/Cas9 began in the lab of Prof. Fabio Fornara (University of Milan), where he used CRISPR/Cas9 to target photoperiod genes of interest in rice and generate mutants that were not previously available. He recently moved to Rothamsted Research, UK, where he works as Genome Editing Specialist, transferring CRISPR/Cas9 technology to hexaploid bread wheat with the aim of improving the efficiency of genome editing in this crop. He is actively involved with AIRIcerca (International Association of Italian Scientists), disseminating and promoting scientific news.