Author: Ansar Ali @Ansar_IPMB, Ph.D. Student at Institute of Plant and Microbial Biology @IPMBsinica, Academia Sinica, Taiwan in @KimmyHo_Lab.
In multicellular organisms, distinct cell types are produced and maintained through the coordination of several progenitor lineages. These lineages behave differently, with some producing a fixed number of progeny while others, like stem cells, act as an endless supply of renewed cells. Cells whose progeny numbers and final identities change in response to changing life circumstances, such as intestinal stem cells that construct organs of various sizes depending on the availability of food (O’Brien et al., 2011), or adult stem-cell lineages where embedded quiescent cells are reactivated in order to maintain and repair tissues, are particularly intriguing from the perspective of flexibility. However, the nature of the stage of these “flexible” cells remains elusive. How is this information stored in the transcriptome? Do these cells’ behaviors reflect their lineage history or their current biological surroundings?
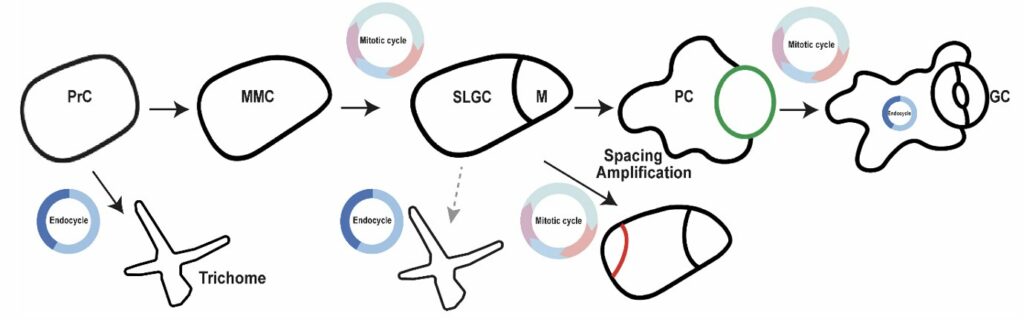
Stomatal lineage ground cells (SLGCs) in the Arabidopsis leaf epidermis exhibit an ideal model to explore developmental flexibilities. They are genetically tractable and experimentally accessible. When a protodermal cell divides asymmetrically to start the stomatal lineage, two distinct daughters are produced: a smaller meristemoid, and a bigger SLGC (Figure 1). These daughters can all divide a finite number of times in different ways. Together, they increase the precursor pool that eventually gives rise to either pavement cells or stomatal guard cells (Figure 1). Although the specific mechanism of SLGCs identity is unknown, hormonal, environmental, or local signals have an impact on SLGCs behavior (Lau et al., 2018; Vatén et al., 2018). For example, cytokinin influences SLGCs divisions to control stomatal generation (Vatén et al., 2018). How SLGCs lose their capacity to divide and change into pavement cells is still a mystery.
The flexibility of division potential and morphological plasticity make SLGCs difficult to isolate
The flexibility of division potential and morphological plasticity make SLGCs difficult to isolate. Recently Ho et al., 2021 used a hybrid strategy to collect SLGCs and their sisters by Fluorescence-Activated Cell Sorting (FACS) coupled with an intensive computational pipeline to generate a transcriptional profile of the enigmatic SLGCs. The SLGCs transcriptional profile named SLGCs cluster consists of 1,016 genes, marked by some well-known SLGCs identity markers such as ERECTA, SPCH, and BASL. GO term analysis showed that SLGCs cluster is enriched in the cell cycle (39.5 %), cytoskeleton organization (14.8 %), and macromolecule methylation (18.0 %), which includes histone methylation, suggesting that SLGCs are division-capable and multipotent. Further analyzing the expression of components involved in the cell cycle progression suggests that SLGCs are at a poised cell state.
By doing a functional study on the SLGC-enriched genes encoding the MYB16 transcription factor and nucleolar localized DEK family members, they also discovered possible roles for transcriptional regulation of SLGC behavior. One of the main transcription factors in the SLGC cluster was MYB16. MYB16p:MYB16-GFP was preferentially expressed in SLGCs in the leaves. SLGCs have a higher expression level of MYB16 than their sister meristemoid. Time-lapse imaging showed that MYB16 degraded more quickly in meristemoids than SLGCs. Additionally, MYB16 expression frequently started after cell division in both daughter cells. This pattern was prominent since the plasma membrane marker slowly bleached out. The phenotypically dominant negative form of myb16SRDX showed decreased stomatal density, suggesting that MYB16 promotes the division of SLGCs. Another highly expressed candidate in the SLGCs cluster was DEK, a putative chromatin-associated protein. Mutant characterization showed a considerable decrease in stomatal density and a tendency toward a lower stomatal index. The overall size of the plant increased, including the pavement cell size. Taken together, these phenotypes indicate that DEK delays differentiation into pavement cell fate and promotes the division of SLGCs.
The authors summarized that SLGCs enriched GO categories shared with other stomata lineage cell types provide a layout for unique gene sets and that SLGCs simultaneously express components in both endocycle and mitotic cell cycle programs. The enriched gene expression is not unique to SLGCs and may find parallels in multipotent precursors of other lineages. Nevertheless, SLGCs may help us understand the stochastic nature of such cell states. A future study using the single cell approach could further help dissect the decision that a SLGC makes – proliferation or differentiation.
Reference
- Ho, C.-M. K., Bringmann, M., Oshima, Y., Mitsuda, N., & Bergmann, D. C. (2021). Transcriptional profiling reveals signatures of latent developmental potential in Arabidopsis stomatal lineage ground cells. PNAS, 118(17), e2021682118. https://doi.org/10.1073/pnas.2021682118/-/DCSupplemental
- Lau, O. S., Song, Z., Zhou, Z., Davies, K. A., Chang, J., Yang, X., Wang, S., Lucyshyn, D., Tay, I. H. Z., Wigge, P. A., & Bergmann, D. C. (2018). Direct Control of SPEECHLESS by PIF4 in the High-Temperature Response of Stomatal Development. Current Biology, 28(8), 1273-1280. https://doi.org/10.1016/j.cub.2018.02.054
- O’Brien, L. E., Soliman, S. S., Li, X., & Bilder, D. (2011). Altered modes of stem cell division drive adaptive intestinal growth. Cell, 147(3), 603–614. https://doi.org/10.1016/j.cell.2011.08.048
- Vatén, A., Soyars, C. L., Tarr, P. T., Nimchuk, Z. L., & Bergmann, D. C. (2018). Modulation of Asymmetric Division Diversity through Cytokinin and SPEECHLESS Regulatory Interactions in the Arabidopsis Stomatal Lineage. Developmental Cell, 47(1), 53-66. https://doi.org/10.1016/j.devcel.2018.08.007
- Zhang, Y., Wang, P., Shao, W., Zhu, J. K., & Dong, J. (2015). The BASL Polarity Protein Controls a MAPK Signaling Feedback Loop in Asymmetric Cell Division. Developmental Cell, 33(2), 136–149. https://doi.org/10.1016/j.devcel.2015.02.022








