Dr Álvaro Montiel Jordá (@spanishbiotech), Postdoctoral researcher at the University of Bristol, writes a commentary on Gobert et al. “Towards plant resistance to viruses using protein-only RNase P.” Nature Communications (2021): 1-6. https://doi.org/10.1038/s41467-021-21338-6
Why is this article important? The authors prove the feasibility of an idea through this proof-of-concept work. As they point out, plant antiviral treatments are usually virus-specific, unfortunately for the moment a ‘broad-spectrum’ plant antiviral does not exist. If they manage to increase the efficiency of a particular protein, named CytoRP, it will become a first-of-its-kind weapon against plant viruses and might start a revolution in agricultural disease management that may lead to a significant reduction in the use of chemicals.
WORLD POPULATION CONTINUES TO GROW
As the world population continues to grow and approaches the 8 billion1, it is of paramount importance to increase agricultural yields to achieve food security. Besides the population growth, other factors, such as crop diseases, threaten food security. Back in 2005, annual losses to world crop production accounted for a total of $550 billion, of which 14.1% (220 billion) were caused by diseases only2. This percentage might even increase with time, as preliminary research suggests that climate change may make plants more susceptible to pathogens3. Given the massive impact of crop diseases on yield, it is no surprise that world pesticide use has been increasing over the last 20 years4, with the associated health and environmental consequences.
New, environmentally friendly technologies are thus needed to protect our crops. New research with Anthony Gobert as first author has come up with a novel strategy to make plants resistant to plant viruses, opening a new door to a reduction in pesticide use in agriculture.
PLANT VIRUSES ARE SPECIAL
Over 80% of plant viruses have an RNA genome. On the contrary, we humans have a DNA genome. Many of these RNA viruses have a particular feature called TLS (tRNA-like structure), which is essential for the virus to replicate and infect its host. TLS are also present in the plant nucleus, mitochondria and chloroplast, where they are cut to allow protein synthesis. This cut is performed by enzymes called PRORP (Protein-Only RNase P). As one would expect, PRORP enzymes are present as well in the nucleus, mitochondria and chloroplasts. Gobert et al., hypothesize that if faced with the virus RNA, plant PRORPs will cut the virus TLS, thus making it impossible for the virus to replicate. However, PRORPs are not present in the cytoplasm, where the virus replicates.
PROPR2 CONTEINS A LOCATION SIGNAL
Proteins contain certain ‘signals’ that tell them where to go in the cell. In the case of PRORP2, which accumulates only in the nucleus, this signal is known as a NLS (Nuclear Localization Signal). The researchers reasoned that by eliminating this signal, the localization of PROPR2 would change and it would no longer be found in the nucleus, but in the cytoplasm instead (Figure 1A). To track the localization of the new PROPR2 protein lacking the NLS (renamed CytoRP), they fused it to a fluorescent protein, visible with the help of a microscope. Indeed, they observed that CytoRP was only present in the cytoplasm of plant cells (Figure 1B).
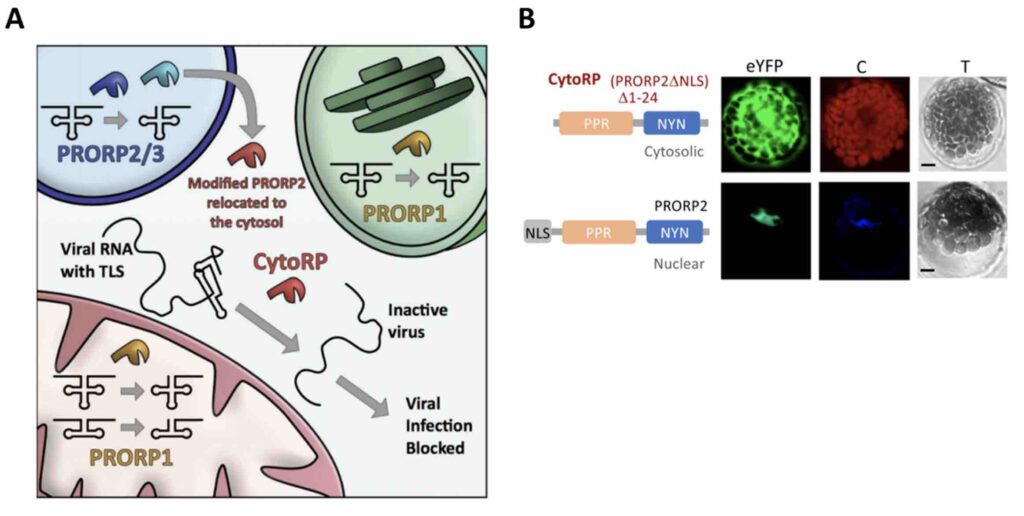
The localization problem seems therefore, solved. Yet, a new question arises. Is CytoRP, the modified version of PRORP2, still able to cut a TLS? To assess that, the authors mixed either CytoRP or PROPR2 with a TLS in vitro (in a test tube) and checked for the presence of cleaved TLS fragments. Both CytoRP and PRORP2 were able to cut the TLS from TYMV (Turnip Yellow Mosaic Virus) and ORMV (Oilseed Rape Mosaic Tobamovirus). However, PRORP2 was more efficient than CytoRP.
Once the two required conditions (ability to cleave the virus RNA and cytoplasmic localization) were proven, the authors sought to test if their engineered CytoRP could work in the real world. In order to do that, they designed different strategies:
- Introduce CytoRP at a level close to that of the naturally present PROPR2 into the plant
- Introduce CytoRP at an artificially very high level and
- Modify the plant genome to turn the endogenous PROPR2 into a CytoRP.
Very importantly, no plants modified through these three strategies showed any detrimental growth effects compared to non-modified ones, demonstrating that the modifications are safe for the plant.
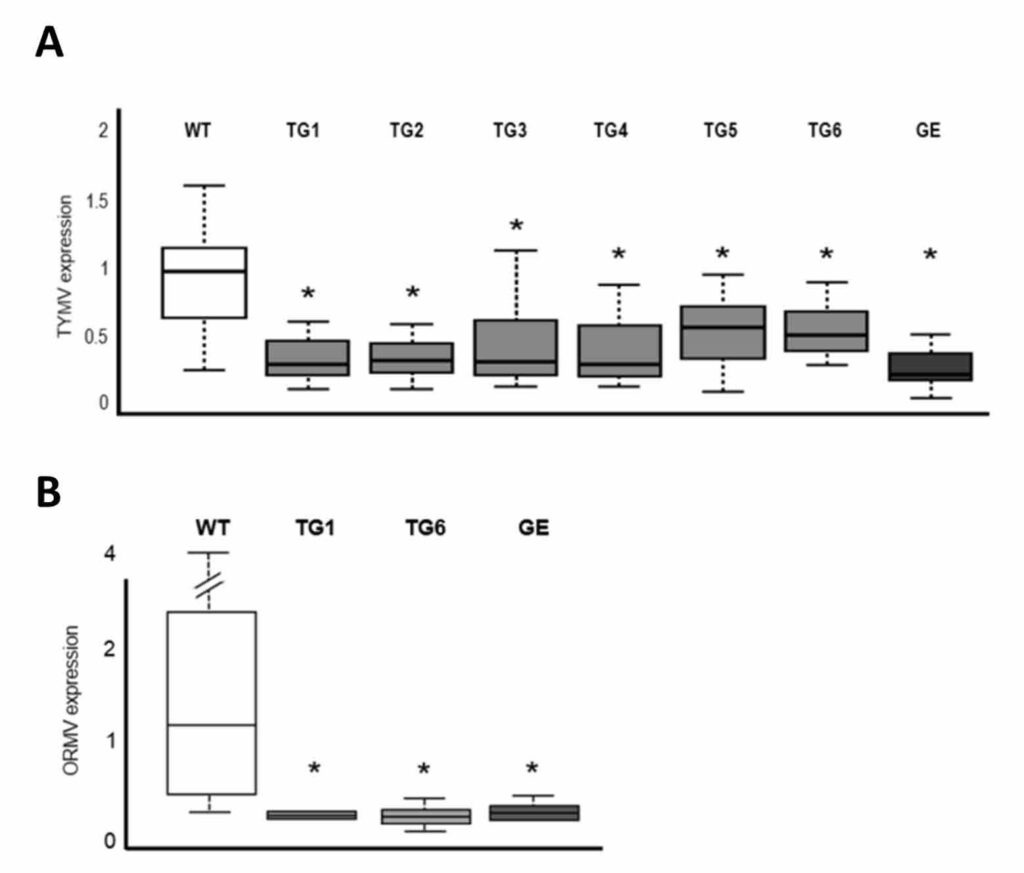
Then, these transgenic plants as well as their wild type (unmodified controls) were inoculated with TYMV and ORMV. As a readout of the infection status, virus load (amount) was analyzed 7 and 28 days after the inoculation. Even if there was no apparent difference when viewed with the naked eye, virus load was significantly lower in plants harboring CytoRP (Figures 2A-B).
However, there was considerable variation between different repetitions of the experiments, which made it impossible to establish a correlation between the amount of CytoRP and the virus accumulation. Further engineering of CytoRP is therefore needed to increase its efficiency.
Read the original paper: Nature Communications
Image credit: Sui-setz / Wikimedia
Bibliography
- https://yaleglobal.yale.edu/content/world-population-2020-overview
- https://ourworldindata.org/pesticides
- Huot, Bethany, et al. “Dual impact of elevated temperature on plant defence and bacterial virulence in Arabidopsis.” Nature communications 8.1 (2017): 1-12.
- Liu, Yongbo, Xubin Pan, and Junsheng Li. “A 1961–2010 record of fertilizer use, pesticide application and cereal yields: a review.” Agronomy for sustainable development 35.1 (2015): 83-93.








