Due to their complexity and microscopic scale, plant-microbe interactions can be quite elusive. Each researcher focuses on a piece of the interaction, and it is hard to find all the pieces let alone assemble them into a comprehensive map to find the hidden treasures within the plant microbiome. This is the purpose of review, to take all the pieces from all the different sources and put them together into something comprehensive that can guide researchers to hidden clues and new associations that unlock the secrets of a system. Like any good treasure map, there are still gaps in the knowledge and the searcher must be clever enough to fill in those gaps to find the “X”. Without a map, there is only aimless wandering, but with a map, there is hope of finding the hidden treasures of the plant microbiome.
Yunzheng Zhang, Nian Wang, and colleagues recently put together a map, “The Citrus Microbiome: From Structure and Function to Microbiome Engineering and Beyond,” which they published in the Phytobiomes Journal. Their map outlines the structure and potential functions of the plant microbiome and how this knowledge can guide us to new engineering feats and a greater understanding of the hidden treasures of the plant microbiome. Once revealed, insights into the microbiome may help researchers grow more resilient citrus plants that are both more suitable for changing climates and more capable of surviving pathogen pressures.
Citrus is a globally important perennial fruit crop that has many economic and emotional ties to society. It is cultivated in more than 140 countries, exposing it to a variety of environmental and disease pressures. Our relationships with citrus are diverse, but not as diverse as the relationship between the citrus and its microbiome. As we shift to an ever-destabilizing environment, it becomes more imperative to understand these interactions to ensure citrus will be present in our lives in the years to come. Nian Wang believes, “harnessing citrus-microbiome interactions to address biotic and abiotic stressors offers an opportunity to increase sustainable citrus production.” To create this treasure map of citrus-microbe interactions, Wang and colleagues established The International Citrus Microbiome Consortium in 2015. Their goal was to form international collaborations to address this global problem. Before understanding the ins and outs of this ecosystem, the team had to know which individual microbes build the citrus microbiome community and how the microbial community may function throughout the different niches of the plant.
After identifying microbes and some of the overall functions the microbiome is performing collectively, the question turns to how they are doing it. Once researchers understand the who, what, and how of the microbiome they can then start to engineer new microbes or design synthetic communities to perform a specific function that may address current deficiencies like crop production, disease spread, and other citrus health ailments. In their review, Zhang, Wang, and colleagues specifically address the first question by combining multiple metagenomic studies into one cohesive paper and confidently stating which microbes make up the citrus microbiome in the rhizosphere (around the root), rhizoplane (on the root), endorhiza (inside the root), and phyllosphere (leaf surface).
This review highlights the exhaustive research that has been done on the citrus microbiome thus far and cleverly assembles this knowledge into one comprehensive figure. The citrus rhizospheres were enriched with microbes belonging to the pyla of Proteobacteria and Bacteroidetes, which is common in other plant rhizospheres. Bradyrhizobium and Burkholderiamicrobes were the most dominant groups in the transition from rhizosphere to rhizoplane. The endorhiza microbiome biomass is approximately a fifth of the rhizosphere biomass and is dominated by Proteobacteria, Firmicutes, and Actinobacteria, as is the phyllosphere.
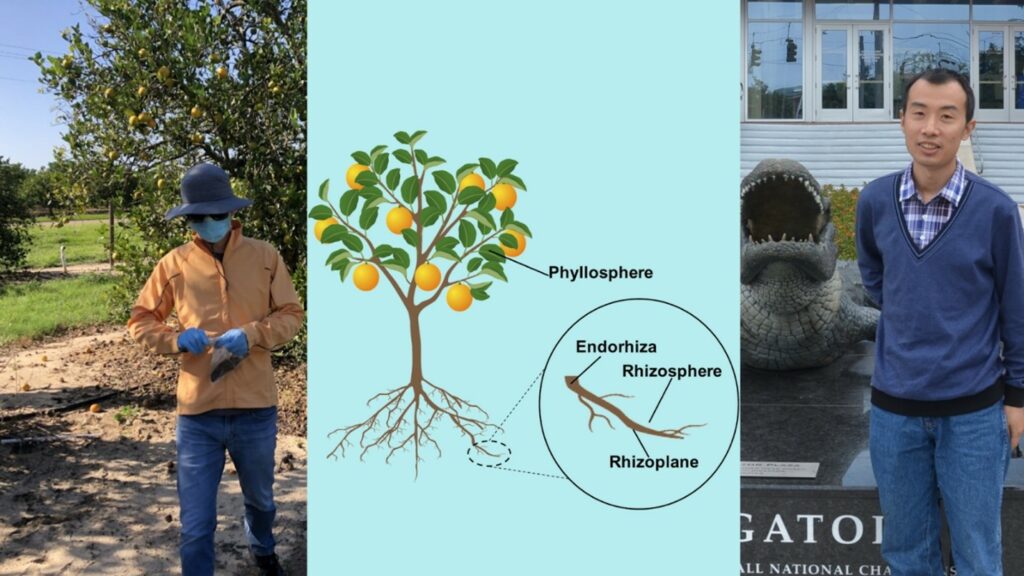
On the left PhD candidate Jin Xu in a citrus orchard; on the right Yunzeng Zhang, who recently became a professor at Yangzhou University in China. In the middle is the first figure of the paper depicting the different compartment of the citrus microbiome. Credit: American Phytopathological Society
But authors note that there is still so much to learn and discover. At this point, researchers only know which microbes are present in some of the tissue, meaning there is still a significant amount of research to be done to unlock the hidden treasures within the citrus microbiome. With this knowledge, it’s time to start incorporating multi-omic technologies, such as transcriptomics (the study of RNA) and metabolomics (the study of metabolites), in addition to the DNA-sequencing techniques that have previously been used. It is an exciting time to be a plant or agricultural scientist because they are on the cusp of integrating novel technologies to understand a world we can’t even see: the world of the microbiome. “I think artificial intelligence will be critical for us to mine a humongous amount of data. Regarding how to utilize the microbiome, I think synthetic community or consortia of microbes, as well as CRISPR-mediated genome editing, will provide the most promising path for the application,” said Wang. An integrated problem of this size, involving so many moving parts, does not only need an international consortium of top researchers but also a highly interdisciplinary team that includes not only plant scientists, microbiologists, and plant pathologists but also experts in the fields of bioinformatics, horticulture, and computer science. Only banded together can this crew of scientists put together the missing pieces of the map and unlock the treasures within the citrus microbiome before it’s too late.
This research article was made possible by the collaboration of Yunzeng Zhang, Pankaj Trivedi, Jin Xu, Caroline Roper, and Nian Wang. Yunzeng Zhang recently started a professorship at Yangzhou University in China. Pankaj Trivedi is now an assistant professor at Colorado State University and Jin Xu is currently a PhD under the mentorship of Dr. Nian Wang. You can find the full article in the Phytobiomes Journal. You can stay up to date with Nian Wang’s Lab’s research on Twitter at @NWangUF.
Read the paper: Phytobiomes Journal
Article source: American Phytopathological Society
Author: Tess Deyett, Ph.D. is a current postdoc under the guidance of Dr. Philippe Rolshausen at the University of California Riverside. She serves as an assistant feature editor for the Phytobiomes Journal. A passionate and driven science communicator, Tess also hosts her own website microbigals.com and podcast The Microbe Moment. She aspires to share how microbes are a constant force of good to every person and that everyone has many unique microbe moments!
Image credit: Free-Photos / Pixabay







