


A nano-sized bio-degradable clay-comprising double stranded ribonucleic acid (dsRNA) could offer a cost-effective, clean and green alternative to chemical-based plant pesticides.
Australian researchers from the University of Queensland have successfully used a gene-silencing spray, named BioClay, a combination of biomolecules and clay, to protect tobacco plants from a virus for 20 days with a single application. Their study has been published in Nature Plants.
“When BioClay is sprayed onto a plant, the virus-specific dsRNA is slowly released from the clay nanosheets into the plant. This activates a pathway in the plant that is a natural defence mechanism. The dsRNA is chopped up into small bits of RNA by enzymes of this pathway. These small bits attack the virus when it infects the plant without altering the plant genome,” explains lead researcher, Neena Mitter.
“Even with current pesticides, we lose up to 40 per cent of our crop productivity because of pests and pathogens. We are hoping that having BioClay in the mix as an environmentally friendly, sustainable crop protection measure will reduce crop losses,” Mitter adds.
“The clay-based delivery technology could represent a positive inflection point in the progress towards commercialisation of topical RNAi. This is a non-GM, environmentally benign and very specific technology.”
John Killmer, APSE
While chemical-based pesticides kill the targeted insect, they can also affect a range of other insects that are beneficial. Mitter says, “BioClay is specific and it only kills the pathogen being targeted. Currently farmers use insecticides to kill the vector that comes with the viruses, but with BioClay we can target the virus itself.”
BioClay field trials may begin in Australia by year-end. “The first test will be on a virus that infects vegetable crops — capsicum, tomato, chilli,” Mitter tells SciDev.Net.
Farmers can use the existing equipment to deliver BioClay and the researchers are hopeful that it will be a commercially viable product for farmers everywhere. The clay component is cheap to make, but not the RNA.
Several companies like APSE, a US based startup, are working on the mass production of RNAs. APSE is developing RNA manufacturing technology for RNA interference (RNAi) or gene silencing applications.
“Our technology for RNA production should be ready in 2-3 years. We are targeting US$2 per gram,” APSE’s John Killmer tells SciDev.Net.
Killmer says, “The clay-based delivery technology could represent a positive inflection point in the progress towards commercialisation of topical RNAi. This is a non-GM, environmentally benign and very specific technology.”
RNAi technology is being used by many in the agriculture industry including the biotech firm Monsanto. The company’s BioDirect technology is focused on applications of RNAi directly onto the leaves of a plant.
Monsanto’s spokesperson John Combest tells SciDev.Net, “As insects develop resistance to certain classes of pesticides, giving farmers another option to control these pests is critical. The idea is not to replace any given system of farming, whether modern GM systems or others — it’s to provide farmers with products that can complement or replace agricultural chemical products.”
This piece was produced by SciDev.Net’s Asia & Pacific desk.
This article was originally published on SciDev.Net. Read the original article.
Established GM technologies are far from perfect
The first genetically modified (GM) crops were approved for commercial use in 1994, and GM crops are now grown on over 180 million hectares across 29 countries. The most used forms of genetic modification are systems that result in herbicide resistance or expression of the Bt toxin in maize and cotton to provide protection against pests such as the European corn borer. These systems both require few novel genes to be introduced to the plant, and allow more efficient use of herbicides and pesticides, both of which are harmful to the environment and human health. Current systems of genetic modification usually involve

Agrobacterium tumefaciens is used to genetically engineer plants in the lab. In nature this bacteria uses its ability to alter plant DNA to cause tumours. Image by Jacinta Lluch Valero used under Creative Commons 2.0.
the use of Agrobacterium vectors, direct transformation by DNA uptake into the plant protoplast, or bombardment with gold particles covered in DNA. However, current systems of transformation are far from perfect. Many beneficial traits such as disease resistance require stacking of multiple genes, something that is difficult with current transformation systems. Furthermore, it is essential that transgenes are positioned correctly within the host genome. Current systems of genetic modification can insert genes into the ‘wrong’ place, disrupting function of endogenous genes or having implications for down or upstream processes. An additional problem is that transfer of transgenes from one line to another requires several generations of backcrossing. However, the past two decades have seen great developments in microbiology. Many new tools and resources are now available that could greatly enhance the biotechnology of the future.
New technologies
Many new and emerging technologies are now available that could transform plant genetic engineering. For example, high throughput sequencing and the wide availability of bioinformatics tools now make identifying target genes and traits easier than ever. Technologies such as site-specific recombination (SSR) and genome editing allow specific regions of the genome to be precisely targeted in order to add or remove genes. Artificial chromosome technology is also part of this emerging group that could be of benefit to plant science. Synthetic chromosomes have already been used in yeast, and widely studied in mammalian systems due to their potential use in gene therapy. Although there have so far been no definitive examples in plants, work has been done in maize that shows the potential of the technology for use in GM crops.
Building an artificial chromosome
A minichromosomes is a small, synthetic chromosome with no genes of its own. It can be programmed to express any desirable DNA sequence that could encode for one, or a number, of genes. An ideal minichromosome would be small and only contain essential elements such as a centromere, telomeres and origin of replication. Once introduced into the plant the minichromosomes should be designed such that interference with host growth and development is minimal. A key requirement is that the chromosome is stable during both meiosis and mitosis. This would ensure introduced genes do not become disrupted or mutated during cell division and reproduction. Gene expression would therefore remain the same for many generations. Finally, the DNA sequence on the minichromosomes could be designed such that it is amenable to SSR or gene editing systems. This would allow re-design and addition of new traits further down the line.
Potential advantages of artificial chromosomes
Plant artificial chromosomes (PACs) have many advantages over traditional transformation systems. For example, to confer complex traits such as disease resistance and tolerance to abiotic stresses such as heat and drought, multiple genes are required. This is not easy with current methods of modification.

PACs could offer a new way to introduce beneficial traits to our crops plants and feed a growing population.Image by Seattle.Romer. Used under Creative Commons 2.0.
However, PACs allow an almost unlimited number of genes to be integrated into the host system. A further possibility that comes from being able to add multiple genes is the addition of new metabolic pathways into the plant. This could allow us to change the nutrients produced by a plant to benefit our diets. Additionally, in a contained environment, plants could be used as a cheap, sustainable way to produce pharmaceuticals. A second major benefit of PACs is that they avoid linkage drag. This is when a desirable gene is closely linked to a deleterious gene that acts to reduce plant fitness. Where this linkage is very tight even repeated backcrossing cannot separate out the genes. Design of new DNA sequences completely avoids this problem, and could allow us to select out detrimental traits from out crop plants.
Regulations for novel biotechnology
Emerging technologies pose new questions to policy makers regarding GM regulation. For example, the use of genome editing, whereby specific sites in the genome are targeted and modified, produces an end product with a phenotype almost identical to one that could be achieved through conventional breeding. This sets genome-edited crops apart from other transgene-containing GM material. For this reason many now argue that genome-edited crops ought not to come under current GM regulations. Much of this argument centres on whether or not to regulate the scientific technique used to produce a crop, or to regulate the end product in the field. For more information on genome editing including current regulations and consensus, see the links at the end of this article.
PACs pose a different set of problems entirely. Minichromosomes would be foreign bodies in the plant, and gene stacking within these introduces even more foreign genes than is possible with current technologies. This would require extensive assessment of both environmental and health effects prior to commercialization. Currently regulatory approval costs around $1-15 million per insertion into the genome. These heavy charges may discourage the further development of minichromosomes technology. However, with PACs it is possible that a particular package of genes could be assessed once, and then transferred into numerous cultivars. This would eliminate the requirement to individually engineer and test every cultivar, so perhaps saving time and money in the long term.
More information on genome editing:
Sense about science genome editing Q & A
The regulatory status of genome-edited crops
The Guardian article on genome editing regulation
A proposed regulatory network for genome edited crops in Nature
A recent workshop on the CRISPR-CAS system of genome editing was held in September 2015 by GARNet and OpenPlant at the John Innes Centre in Norwich, UK. You can read the full meeting report here.
Following on from last week’s post, Now That’s What I Call Plant Science 2015, we bring you a year in Plant Science!

Image credit: Jean Weber. Used under license CC BY 2.0.
The year began with a surprising paper that turned our understanding of the phytohormone auxin on its head. Researchers in China and the USA created Arabidopsis knockout mutants of AUXIN BINDING PROTEIN 1 (ABP1), expecting them to fail to respond to auxin and have developmental defects, as previously seen in the abp1-1 knockdown mutant. Instead, these plants were indistinguishable from wild type plants, leading the authors to conclude that ABP1 is not required for auxin signaling or Arabidopsis development as previously believed.
Read the paper in PNAS: Auxin binding protein 1 (ABP1) is not required for either auxin signaling or Arabidopsis development.
A paper later in the year from the same authors found that the embryonic lethality of the abp1-1 mutant is actually caused by the off-target linked deletion of the adjacent BSM gene.
Read this paper in Nature Plants: Embryonic lethality of Arabidopsis abp1-1 is caused by deletion of the adjacent BSM gene.
The tale of ABP1 was examined in more detail on the GARNet blog, Weeding the Gems, which concluded: “In many ways this story is an excellent example of how science should work, where claims are independently tested to ensure that earlier experiments have been conducted or interpreted correctly.” Click here to read more.
A clever experiment from Germany led to a significant breakthrough in crop protection from insect pests.
When double-stranded RNA (dsRNA) is present within a eukaryotic cell, it is cleaved by the Dicer enzyme to form short interfering RNAs. These can bind to complementary RNA within a cell to target it for destruction, thus silencing the corresponding gene expression. This process is known as RNA interference (RNAi).
RNAi has previously been used to tackle insect herbivory by expressing insect-specific dsRNA in plants; however the protection has previously been incomplete. In this new study, published in Science, researchers produced dsRNA within chloroplasts, which do not have RNAi machinery. When dsRNA is expressed in the cytoplasm, the plant’s own Dicer enzyme breaks most of it down. When expressed in the chloroplasts, the dsRNA remained intact when eaten by insects, which proved much more effective at killing these pests.
Read the paper here: Full crop protection from an insect pest by expression of long double-stranded RNAs in plastids.
Another crop protection study followed in March, when researchers in China cloned the genetic locus in rice that confers broad-spectrum resistance to planthoppers – insect pests that cause the loss of billions of dollars of crops per year. Three lectin receptor kinase genes were found in rice cultivars from the Philippines, which enable plants to survive an infestation of insects. When cloned into a susceptible rice cultivar, these genes conferred resistance to two different planthopper species.
Understanding the genetic basis of resistance is very important as marker-assisted breeding and selection could be used to develop resistant rice varieties, and potentially utilized in other species of cereal.
Read the paper in Nature Biotechnology: A gene cluster encoding lectin receptor kinases confers broad-spectrum and durable insect resistance in rice
A European collaboration led to the development of 3DCellAtlas, a computational approach that semi-automatically identifies cell types in a developing 3D organ without the need for transgenic lineage markers. This program will enable the interpretation of dynamic organ growth and the spatial and temporal context of developmental cell divisions that produce the resultant plant. It could be integrated with growth in different conditions or with developmental mutants to examine exactly how these processes affect growth in 3D.
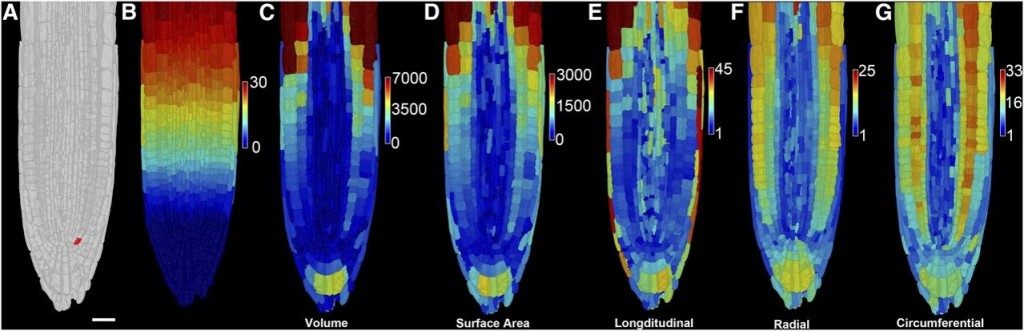
Image credit: Montenegro-Johnson et al., 2015. Digital Single-Cell Analysis of Plant Organ Development Using 3DCellAtlas. The Plant Cell, vol. 27 no. 4, 1018–1033.
A special issue of the Plant Biotechnology Journal was published in May, focusing on the amazing advances in molecular farming. While the entire issue is worth delving into, we were particularly intrigued by the review on moss-made pharmaceuticals, which outlines the rapid progress made in the field.
The model moss Physcomitrella patens has rapidly become one of the organisms of choice in biotechnology, with a fully sequenced genome and an outstanding toolbox for genome-engineering. The authors describe how moss-made pharmaceuticals can easily be produced while remaining remarkably more stable from batch to batch than cultured animal cells. The system is easily scalable, making their production highly cost effective, and safe. The first moss-made pharmaceuticals are currently in clinical trials, so keep an eye out for much more from this field over the next few years.
Read the review: Moss-made pharmaceuticals: from bench to bedside.
In June, US researchers discovered a new role for chloroplast stromules, protrusions that extend from the surface of all plastid types. The function of stromules has been difficult to determine, but this research, published in Developmental Cell, suggests that they may provide a mechanism by which plastid signals are conveyed to the nucleus. The paper shows that chloroplast stromules are induced by defense responses such as programmed cell death signaling, and that the stromules extend to form dynamic connections with the nucleus. The stromules may therefore aid in the amplification and/or transport of immune response signals into the nucleus.
Read the paper: Chloroplast Stromules Function during Innate Immunity.
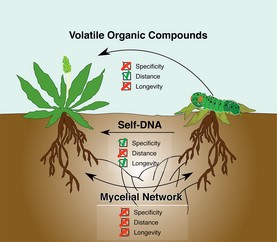
Image credit: Veresoglou et al., 2015. Self-DNA: a blessing in disguise? New Phytologist, vol. 207, no. 3, 488–490.
In late 2014 and early 2015, Italian researchers published a set of articles showing that extracellular self-DNA, DNA from conspecifics, could inhibit the growth of organisms from a wide range of taxa, including plants, bacteria, fungi and animals. Conversely, these organisms were not affected by extracellular DNA from other unrelated species.
In July, New Phytologist published a letter offering an interpretation of the data as it relates to plants. Plants could interpret extracellular self-DNA as an indicator of intraspecific competition (which seeds could use as a cue to remain dormant) or of a hostile environment that has already caused the death of conspecifics, signaling them to ramp up their pre-emptive immune response to increase survival after neighbors have been damaged or killed. There are still a lot of mechanisms and ecological effects to be investigated in this new field, but this letter suggests several interesting avenues to investigate.
Read the article: Self-DNA: a blessing in disguise?
Original research papers in New Phytologist:
Inhibitory effects of extracellular self-DNA: a general biological process?
A US study in August revealed a surprising degree of conservation in gene expression patterns across a wide range of plant taxa during root development. This was particularly interesting because the spikemoss Selaginella was shown to use many of the same genes as the evolutionarily distant angiosperms, despite the fossil record suggesting that roots evolved independently in these two lineages. Perhaps roots in these two groups evolved by independently recruiting the same developmental program, or perhaps by elaborating on a previously unknown proto-root that existed in the common ancestor of vascular plants.
Read the paper in The Plant Cell: Conserved Gene Expression Programs in Developing Roots from Diverse Plants.
Salt stress can significantly reduce the growth and yield of plants. Researchers in Germany identified two components of the cellulose synthase complex that directly interact with the microtubules and promote their dynamics, which interestingly were highly produced during salt stress conditions. During salt stress, cellulose microtubules depolymerize, however the newly discovered compounds, known as Companions of Cellulose Synthase, promote the reassembly of the microtubule to allow cellulose synthesis to continue.
Read the paper in Cell: A Mechanism for Sustained Cellulose Synthesis during Salt Stress
Throughout the year the GM debate in Europe reached several important milestones. In January the European Union (EU) changed its rules, giving individual countries more flexibility to decide for themselves whether or not to plant GM crops. In February, the UK Science and Technology Committee report stated that EU regulations preventing GM crops are not fit for purpose, and that they should be replaced with a trait-based system.
In October, EU member states revealed their stances on GM crops, with over half of Europe opting out of growing GM crops. Germany was the largest country to opt out of growing GM. The full list can be viewed here: Restrictions of geographical scope of GMO.
Read the news articles here:
EU changes rules on GM crop cultivation – January 2015
EU regulation on GM Organisms not ‘fit for purpose’ – February 2015
Half of Europe opts out of new GM crop scheme – October 2015

Image credit: University of Warwick, UK.
A collaboration between South African and UK scientists revealed how plants can use their circadian clock to pre-emptively boost their immune resistance at dawn, when fungal infection is most likely. Plants tend to decrease in susceptibility at dawn, but those with dysfunctional circadian clocks remained highly susceptible throughout the day. The research also showed that jasmonate signaling plays a crucial role in the circadian timing of resistance.
Read the article in The Plant Journal: Jasmonate signalling drives time-of-day differences in susceptibility of Arabidopsis to the fungal pathogen Botrytis cinerea.

Image credit: Guo & Liu., 2015. A single-nucleotide exon found in Arabidopsis. Scientific Reports, 5:18087.
Researchers in China published the surprising finding that a single-nucleotide exon exists in the APC11 gene in Arabidopsis. This is the smallest exon ever to be discovered before. The team used an elegant set of APC11-GFP constructs to show that intron splicing around the single-nucleotide exon is effective in both Arabidopsis and rice. This finding has implications for future genome annotations, which might reveal many more single-nucleotide exons.
Read the paper in Scientific Reports: A single-nucleotide exon found in Arabidopsis.
What a wonderful year of science! What new knowledge will 2016 bring?