Researchers identify a unique version of a photosynthetic enzyme that has been in use for billions of years.
A team of scientists has discovered an ancient form of rubisco, the most abundant enzyme on Earth and critical to life as we know it.
Found in previously unknown environmental microbes, the newly identified rubisco provides insight into the evolution of the photosynthetic organisms that underlie the planet’s food chains.
“Rubisco is the primary driver for producing food, so it can take CO2 from the atmosphere and fix that into sugar for plants and other photosynthetic organisms to use,” said Doug Banda, a postdoctoral scholar in the lab of Patrick Shih, a UC Davis assistant professor and the director of Plant Biosystems Design at the Joint BioEnergy Institute (JBEI), which is managed by Lawrence Berkeley National Laboratory (Berkeley Lab). “It is also one of the oldest carbon-fixing enzymes on the planet.”
Form I rubisco, which is found in plants, algae, and cyanobacteria, has a deep evolutionary history with the planet, going back nearly 2.4 billion years to the Great Oxygenation Event, when cyanobacteria literally transformed the Earth’s atmosphere by introducing oxygen to it through photosynthesis. Rubisco’s role in this foundational event makes it a key focus of scientists studying the evolution of life, as well as scientists seeking to develop bio-based fuels and renewable energy technologies.
In a study appearing in Nature Plants, Banda and researchers from UC Davis, UC Berkeley, and Berkeley Lab report the discovery and characterization of a previously undescribed lineage of form I rubisco – one that the researchers suspect diverged from form I rubisco prior to the evolution of cyanobacteria.
Found through metagenomic analysis of environmental samples and synthesized in a lab, the new lineage, called form I’ rubisco, gives researchers new insights into the structural evolution of form I rubisco, potentially providing clues as to how this enzyme changed the planet.
“This could’ve been what a rubisco looked like before the rise of oxygen more than 2.4 billion years ago,” said Shih, noting that the form I’ rubisco provides scientists with a window into how ancient microbes might’ve fixed carbon before the rise of cyanobacteria and the form I rubisco.
An invisible world
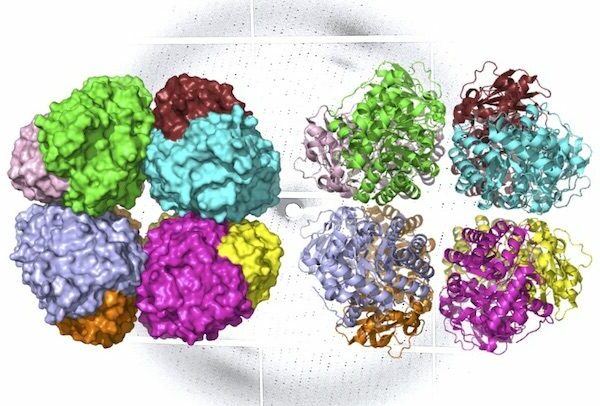
Form I rubisco is a hexadecamer, meaning it’s built from eight core, large molecular subunits with eight small subunits perched on top and bottom. Each piece of this protein’s structure is integral to photosynthesis, and thus the carbon fixation process.
Other functional forms of rubisco exist in bacteria and microorganisms of the Archaea domain. These variants come in different shapes and sizes, and all perform the same step of photosynthesis. However, form I rubisco is responsible for the vast majority of carbon fixation on Earth.
Study co-author and collaborator Professor Jill Banfield, of UC Berkeley’s Earth and Planetary Sciences Department, uncovered form I’ rubisco after performing metagenomic analyses on groundwater samples. Metagenomic analyses allow researchers to examine genes and genetic sequences from uncultured microorganisms found in the environment.
Using the genes and genetic sequences provided by Banfield, Banda, and Shih successfully expressed form I’ rubisco in the lab using E. coli. To learn how this newly identified form functions and how it compares to previously discovered rubisco enzymes, the scientists needed to build precise, 3D models of its structure. For this task, the lead authors turned to Berkeley Lab structural biologists Paul Adams, Henrique Pereira, and Michal Hammel.
First, Adams and Pereira performed X-ray crystallography – an approach that can generate images of molecules with atomic-level resolution – at Berkeley Lab’s Advanced Light Source (ALS). Then, to capture how the enzyme’s structure changes during different states of activity, Hammel applied a technique called small-angle X-ray scattering (SAXS) using the SIBYLS beamline at the ALS.
SAXS is a lower-resolution technique, but unlike crystallography – which requires that sample molecules are frozen in crystal form – SAXS is performed in solution. When the data from the two approaches are combined, scientists can construct unprecedented models of complex molecules as they appear in nature.
“Like many enzymes key to life, rubisco has several protein domains connected together, and as it binds with other molecules during the photosynthesis reaction, it will cycle through different arrangements of those domains,” said Hammel, a biophysicist in Berkeley Lab’s Molecular Biophysics and Integrated Bioimaging (MBIB) Division. “Our techniques really worked hand-in-hand to reveal how this new, novel rubisco behaves in real-world, physiological conditions.”
The ALS investigations showed that like form I rubisco, form I’ rubisco is built from eight large subunits. However, it doesn’t possess the small subunits that were previously thought to be essential to its carbon-fixing function.
The researchers now believe that form I’ rubisco represents a missing link in the evolutionary history of form I rubisco’s structure.
“The discovery of an octameric rubisco that forms without small subunits allows us to ask [evolutionary] questions about what life would’ve looked like without the functionality imparted by small subunits,” said Banda.
Following the success of the structural investigation into form I’ rubisco, Shih has enlisted Hammel, Adams, and Pereira to apply their complementary approach for studies of other crucial plant enzymes, including additional forms of rubisco.
“We’ve been working together at Berkeley Lab for over 10 years now, and it was really satisfying to be able to see what crystallography and SAXS combined can do to understand biology problems,” said Pereira, an MBIB biophysicist. “Once, the scientists who use these different structural biology techniques would have seen themselves as in competition, racing each other to solve structures. But now it’s pure collaboration.”
The ALS is a Department of Energy (DOE) user facility and JBEI is a DOE Bioenergy Research Center. The crystallography beamline used in this research is operated by the Berkeley Center for Structural Biology and funded by the Howard Hughes Medical Institute. The SIBYLS beamline is supported by the National Cancer Institute grant Structural Biology of DNA Repair and the DOE Office of Science. This work was supported in part by the DOE Office of Science.
Read the paper: Nature Plants
Article source: Berkeley Lab
Author: Aliyah Kovner
Image: A ribbon diagram (R) and molecular surface representation (L) of carbon-fixing form I’ rubisco, showing eight molecular subunits without the small subunits. An x-ray diffraction pattern of the enzyme, also generated by the research team, is in the background. Credit: Henrique Pereira/Berkeley Lab.