For life on Earth, it is essential that plants carry out photosynthesis and ultimately produce oxygen and chemical energy with the help of sunlight. Researchers from Göttingen and Hannover have now succeeded for the first time in visualising the copying machine of chloroplasts, the RNA polymerase PEP, in high-resolution 3D. The detailed structure provides new insights into the function and evolution of this complex cellular machine, which plays a central role in reading the genetic instructions for photosynthesis proteins.
Without photosynthesis, there would be no air to breathe – it is the basis of all life on Earth. This complex process allows plants to convert carbon dioxide and water into chemical energy and oxygen using light energy from the sun. The conversion takes place in the chloroplasts, the heart of photosynthesis. Chloroplasts developed in the course of evolution when the ancestors of today’s plant cells absorbed a photosynthetic cyanobacterium. Over time, the bacterium became increasingly dependent on its “host cell”, but maintained some significant functions such as photosynthesis and parts of the bacterial genome. The chloroplast therefore still has its own DNA, which contains the blueprints for crucial proteins of the “photosynthesis machinery”.
With PEP to energy
“A unique molecular copying machine, an RNA polymerase called PEP, reads the genetic instructions from the chloroplasts’ genetic material,” explains Prof. Dr. Hauke Hillen, research group leader at the Max Planck Institute (MPI) for Multidisciplinary Sciences, professor at the University Medical Center Göttingen and member of the Göttingen Cluster of Excellence “Multiscale Bioimaging” (MBExC). It is essential for activating the genes required for photosynthesis, Hillen emphasises. Without a functioning PEP, plants cannot photosynthesise and remain white instead of turning green.
Not only the copying process is complex, but also the copying machine itself: It consists of a multi-subunit core complex, whose protein parts are encoded in the chloroplast genome, and at least twelve associated proteins, called PAPs. The nuclear genome of the plant cell provides the blueprints for these. “So far, we have been able to characterise some individual parts of the chloroplast copying machine structurally and biochemically, but we lacked a precise insight into its overall structure and the functions of the individual PAPs,” says Prof. Dr. Thomas Pfannschmidt, professor at the Institute of Botany at Leibniz University Hannover.
Detailed snapshot in 3D
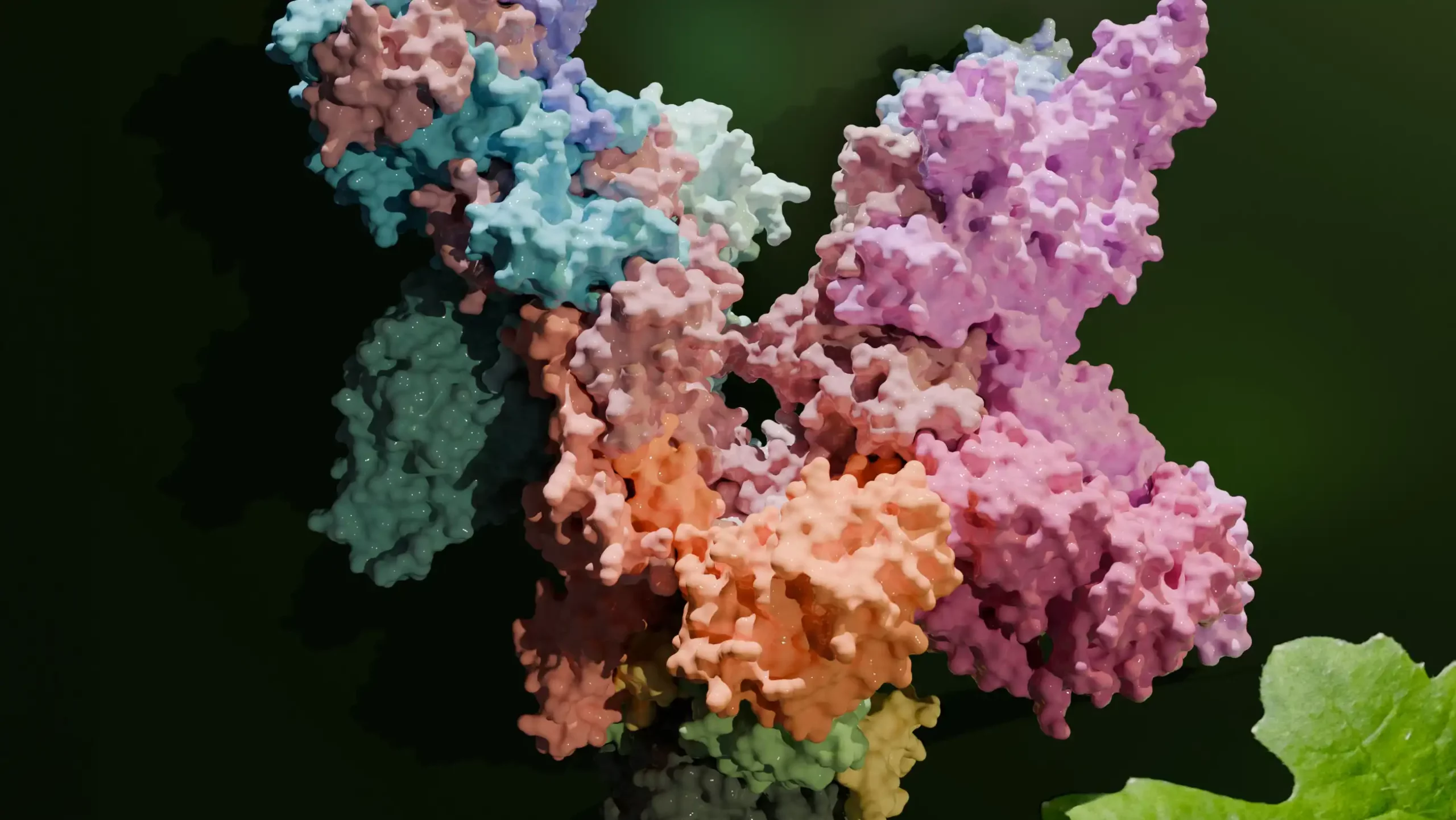
In close collaboration, researchers led by Hauke Hillen and Thomas Pfannschmidt have now succeeded for the first time in visualising a 19-subunit PEP complex in 3D at a resolution of 3.5 angstroms – 35 million times smaller than a millimetre.
“We isolated intact PEPs from white mustard, a typical model plant in plant research,” describes Frederik Ahrens, a member in Pfannschmidt’s team and one of the first authors of the study now published in the journal Molecular Cell. Using cryo-electron microscopy, the scientists then created a detailed 3D model of the 19-part PEP complex. For that, the samples were flash-frozen ultra-fast. The researchers then photographed the copying machine thousands of times and down to the atomic level from numerous angles and combined them into an overall image using complicated computer calculations.
“The structural snapshot showed that the PEP core is similar to those in other RNA polymerases, such as in bacteria or the cell nucleus of higher cells. However, it contains chloroplast-specific features that mediate the interactions with the PAPs. The latter we find only in plants and they are unique in their structure,” explains Paula Favoretti Vital do Prado, PhD student at the MPI, member of the MBExC’s Hertha Sponer College, and also first author of the study. Scientists had already assumed that the PAPs fulfil individual functions in reading the photosynthesis genes. “As we could show, the proteins arrange themselves in a special way around the RNA polymerase core. Based on their structure, it is likely that the PAPs interact with the core complex in various ways and are involved in the gene reading process,” Hillen adds.
Understanding the evolution of photosynthesis
The research collaboration also used databases to search for evolutionary clues. They wanted to find out whether the observed architecture of the copying machine is similar in other plants. “Our results indicate that the structure of the PEP complex is the same in all land plants,” says Pfannschmidt. The new findings on the copying process of chloroplast DNA help us better understand the fundamental mechanisms of the photosynthesis machinery’s biogenesis. They might also be valuable for biotechnological applications in the future.
The study was funded by the German Research Foundation (FOR2848, SFB1565, PF323-7 and SPP 2237 MadLand (PF323-9)) and within the framework of the Excellence Strategy (EXC 2067/1 – 390729940) via the Cluster of Excellence “Multiscale Bioimaging: From Molecular Machines to Networks of Excitable Cells” (MBExC) as well as by the European Research Council (ERC) within the framework of the EU Horizon 2020 programme with the ERC Starting Grant MitoRNA (Grant agreement no. 101116869).
Read the paper: Molecular Cell
Article source: Leibniz University Hannover
Image: The image shows a high-resolution 3D model of the plant RNA polymerase PEP, which plays a central role in photosynthesis. Credit: Paula Favoretti Vital do Prado and Johannes Pauly / MPI-NAT, UMG