A team of biologists has identified that the pathogenic fungus Verticillium dahliae, responsible for wilt disease in many crops, secretes an ‘effector’ molecule to target the microbiome of plants to promote infection. The research was performed by the team of Alexander von Humboldt Professor Dr Bart Thomma at the University of Cologne (UoC) within the framework of the Cluster of Excellence on Plant Sciences (CEPLAS) in collaboration with the team of Dr Michael Seidl at the Theoretical Biology & Bioinformatics group of Utrecht University in the Netherlands. The study ‘An ancient antimicrobial protein co-opted by a fungal plant pathogen for in planta mycobiome manipulation’ has appeared in the Proceedings of the National Academy of Sciences (PNAS).
Scientists increasingly recognize that an organism’s microbiome – the totality of bacteria and other microbes living in and on it – is an important component of its health. For humans as well as other animals, particular microbes inhabiting the gut and the skin have beneficial effects. This is similarly true for plants. Moreover, it has been established that plants can ‘recruit’ beneficial microbes from their environment, for instance from the soil surrounding the roots, to help them withstand disease.
At the UoC, together with Dr Nick Snelders lead author of the study Dr Thomma hypothesized that if plants can do this, perhaps some pathogens have ‘learned’ to perturb this ‘cry for help’ and disturb the plant’s microbiome in order to promote invasion. Thus, in addition to the direct suppression of the plant host’s immune responses, these pathogens can suppress immunity indirectly by affecting the plant’s healthy microbiome.
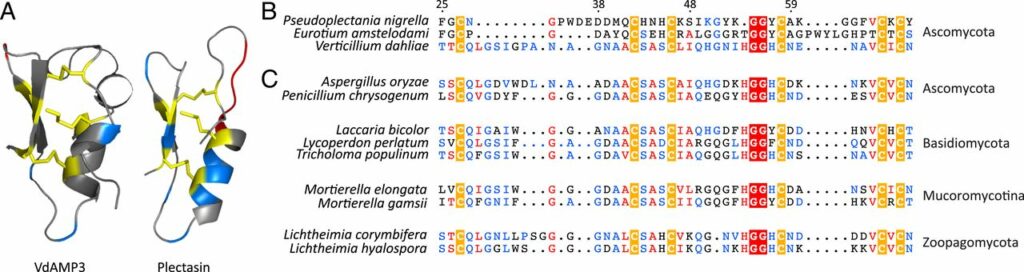
Verticillium dahliae is a well-known pathogen of many plants, including greenhouse crops like tomatoes and lettuce, but also olive trees, ornamental trees and flowers, cotton, potatoes, and others. The current study shows that the fungus secretes the antimicrobial protein VdAMP3 in order to manipulate the plant’s microbiome as an effector.
Generally, effector molecules target components of the host immune system, leading to immune suppression. The authors have now shown that these targets extend to inhabitants of the host’s microbiome: during host colonization, the VdAMP3 molecule suppresses beneficial organisms in the microbiome of the plant, leading to microbiome disturbance or ‘dysbiosis’, so that the fungus can complete its life cycle and produce progeny that can spread and start new infections.
‘In terms of evolution, the molecule that is secreted is very old. Homologs also occur in organisms that are not pathogenic on plants,’ said Thomma. ‘It looks like Verticillium “used” the molecule to “exploit” it during the process of disease development on the host. Interestingly, the molecule does not act like a broad-spectrum antibiotic that targets any microbe, but specifically against “competitor” fungi that have abilities to hinder Verticillium.’
Initially, the researchers wanted to find out whether Verticilium had ‘effectors’ in its arsenal that have antimicrobial activity. Different candidates were then tested for their ability to inhibit the growth of test microbes in the laboratory. VdAMP3 was one of these potential candidates. Follow-up experiments then showed that without the ‘effector’, other fungi – antagonists – could thrive and suppress the formation of Verticillium reproduction structures. VdAMP3 is mainly active in later stages of disease development, when Verticillium needs to produce these new reproduction structures to start infections of new hosts. However, this is not the first such molecule the scientists have identified: A year ago, they found a molecule that does not act against competing fungi, like VdAMP3, but against bacterial competitors.
‘Together, these findings demonstrate that pathogens use various molecules at various stages of the disease process to manipulate the healthy microbiome of a host to cause disease. This shows that it is important to look beyond the “binary interaction” between a pathogen and a host if we want to understand disease. Rather, we must take the entire microbiome of the host into account as well, looking at the host as a “holobiont” – the ecological unit formed by the host and all the organisms living in and on it,’ Thomma added.
In the long term, a better understanding of these mechanisms will help to develop more resilient plants and better strategies for crop protection. In the face of a growing world population, limited farmland, and the need to reduce environmental impact and pollution, one of the major aims of plant scientists is to increase the yield of field crops while minimizing our ecological footprint on the environment. ‘Learning more about these effector molecules that help pathogenic fungi infect crop plants may lead to new ways to safeguard against them,’ said Snelders.
In future studies, the authors aim to find further effector proteins with selective antimicrobial activity – from Verticillium, but also from other pathogens that have other infection strategies. ‘Unravelling how these molecules work, and how they can inhibit the one microbe while not affecting the other is important to discover novel mechanisms to target microbes, which may ultimately even lead to the development of novel antibiotics,’ Snelders concluded.
The Cluster of Excellence on Plant Sciences (CEPLAS) is a joint initiative of Heinrich Heine University Düsseldorf (HHU), the University of Cologne (UoC), the Max Planck Institute for Plant Breeding Research Cologne (MPIPZ), and Forschungszentrum Jülich (FZJ). In CEPLAS, scientists are developing innovative strategies for sustainable plant production. With generous funding by the Alexander von Humboldt foundation through an Alexander von Humboldt professorship, Thomma’s recently re-located working group from Wageningen University in the Netherlands to the UoC focuses on identifying mechanisms that underlie the pathogenicity of fungi on plant hosts.
The Theoretical Biology & Bioinformatics group of Utrecht University, the Netherlands, uses computational biology, bioinformatics, modelling, and big data to address both fundamental and applied questions in the life sciences. Dr Seidl’s Fungal Evolutionary Genomics group focuses on genome evolution and function to understand how fungi evolve and adapt to novel or altered environments, both on short and long evolutionary timescales.
Read the paper: PNAS
Article source: University of Cologne
Image credit: Charles Andres / Wikimedia








