Scientists at the Leibniz Institute of Plant Biochemistry (IPB) have succeeded for the first time in stably and precisely inserting large gene segments into the DNA of higher plants very efficiently. To do this, they optimized the gene-editing method CRISPR/Cas, commonly known as “genetic scissors.”
The improved CRISPR method offers great opportunities for the targeted modification of genes in higher plants, both for breeding and research. The study, led by Prof. Alain Tissier and Dr. Tom Schreiber, has been published in Molecular Plant.
CRISPR/Cas is a method with enormous potential for the targeted modification of individual genes. However, this does not apply to all kinds of genetic modifications that breeders and scientists have on their wish lists. While the genetic scissors are ideal for knocking out genes, i.e., switching off or removing existing genes, they do not work well for precisely inserting genes or replacing gene segments. To date, genetic scissors have been too inefficient and therefore of little use for the targeted insertion of genes into the DNA of higher plants.
“The reason for this is the plant’s internal repair machinery for DNA breaks,” says Schreiber. These repair enzymes are immediately present as soon as damage to the DNA occurs. They also recognize the smooth cuts made by the genetic scissors and instantly rejoin the two severed DNA strands of the double helix. This gluing together of the cut DNA occurs very quickly and not very precisely; there are minor losses of information in which tiny sections of DNA are lost or added.
“These inaccuracies are not a problem in knock-out projects and are even desirable,” says Schreiber, “because I want to switch off the gene anyway. But if I want to insert a gene, it has to be done very precisely. The genetic information must be inserted exactly, not a single component may be missing and not a single additional component may be integrated, otherwise the gene loses its function and the entire experiment was in vain.”
For this reason, CRISPR/Cas-mediated precise and scar-free insertion of larger genes or DNA segments has only been successful in rare individual cases to date. In order to increase the success rate of gene insertion, the Halle scientists equipped the genetic scissors with an additional enzyme, a so-called exonuclease.
Exonucleases can alter the DNA cleavage sites created by the genetic scissors in such a way that the cell’s internal repair enzymes can no longer recognize and mend the DNA damage. The DNA segment to be inserted by CRISPR/Cas would therefore have enough time to integrate itself at the correct position through another, very precise, cellular repair mechanism.
In the experiment, the Halle scientists tested various exonucleases of viral, bacterial, plant, and human origin for their ability to increase the number of precise gene insertion events. They introduced the genetic scissors with the corresponding exonucleases and a gene X segment into the leaf cells of the tobacco plant Nicotiana benthamiana.
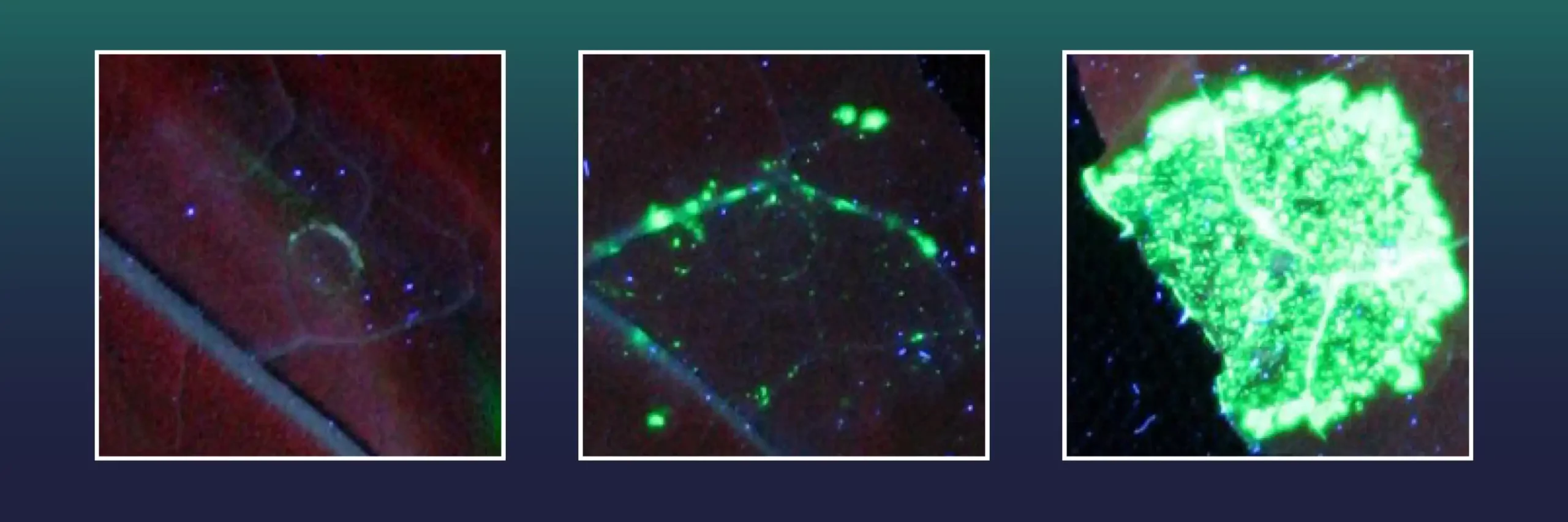
These tobacco cells had previously been equipped with a gene for a green fluorescent marker. They also contained a destroyed gene X, which is required for the formation of the green fluorescent dye. However, the fluorescent marker cannot be generated as long as a large part of gene X’s genetic information is missing.
The green marker can only be produced when the missing gene section of X is precisely reinserted using CRISPR/Cas, thus repairing gene X. Each cell with successful gene insertion will then fluoresce in green and researchers can simply count the rate of successful gene insertion events.
Two of the exonucleases tested, including one from the herpes virus family, proved to be particularly effective. Using these, the team from Halle achieved 38 times more perfect gene insertion events than with CRISPR/Cas alone.
This experimental approach was then tested with other genes to be incorporated and in other plants, namely thale cress (Arabidopsis thaliana) and wheat. Since the gene insertion in the tobacco plants took place only locally in the leaves, the integrated gene was lost in the next daughter generation and was therefore only present in the genome for a limited time.
This is why in Arabidopsis and wheat, the Halle CRISPR experts tried to incorporate the gene into germline cells to ensure a stable inheritance to future plant generations. With the help of the tested exonucleases, the stable, i.e., heritable, knock-in of genes turned out successful in Arabidopsis with a tenfold increase in frequency and in wheat in more than 1% of the daughter plants.
“One percent doesn’t sound like much at first,” Schreiber explains, “but if a breeder wants to introduce a certain trait into his plant, he would only have to screen around 50–100 first-generation daughter plants using our optimized CRISPR/Cas method to find a plant with the desired trait. This would save a considerable amount of time compared to conventional breeding methods, where 500 to 1,000 plants would have to be analyzed for this purpose.”
Therefore, the optimized CRISPR/Cas method is a promising tool for the targeted insertion of genes into higher plants and possibly also into other organisms. In the future, plant breeders could use this method, for example, to reintroduce lost resistance genes against pathogens from wild species or old cultivated varieties into modern, high-yielding elite varieties. That way, desirable traits like these could enhance plant breeding and contribute to the development of more robust crop varieties.
For science, this approach offers great opportunities to elegantly replace certain plant genes with modified copies of themselves in a single step. This is particularly helpful in elucidating gene function.
Read the paper: Molecular Plant
Article source: Leibniz Institute of Plant Biochemistry (IPB)
Author: Sylvia Pieplow
Image: Each green fluorescent spot represents a leaf cell in which a large gene segment was successfully inserted into the plant’s genome. Different variants of Cas9 were used: an inactiv Cas9 (left), an active Cas9 (center) and Cas9 fused with an exonuclease (right). Credit: Tom Schreiber, IPB, Leibniz Institute of Plant Biochemistry, IPB.