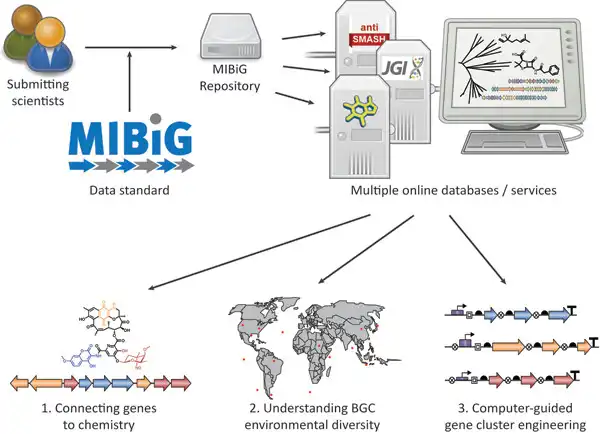
The Minimum Information about a Biosynthetic Gene cluster (MIBiG), first stablished in 2015, was recently updated. The MIBiG specification provides a robust community standard for annotations and metadata on biosynthetic gene clusters and their molecular products.
The MIBiG database (available online) is a curated collection of Biosynthetic Gene cluster annotations that adhere to the MIBiG standard. It serves as a valuable resource for researchers working on natural products. The MIBiG database gathers currently genomic and molecular data from nearly 2500 biosynthetic pathways of fungal, bacterial, and plant secondary metabolites.
An international consortium of 288 scientists from nearly 180 research institutions and companies across 33 countries participated in an annotation and curation effort of MIBiG. During this process, the structure of this open access online database (https://mibig.secondarymetabolites.org/) was revised. In addition to manual verification of the database entries, new information was added, such as biological activities, biosynthesis steps, and the nature of the evidence linking a biosynthetic gene cluster (BGC) to secondary metabolites.
The description of these updates has just been published in the journal Nucleic Acids Research.
Read the paper: Nucleic Acids Research
Article source: SPS Network, Bioger
Author: JFD, adapted.
Image credit: MIBiG database






