Accelerating plant evolution with CRISPR paves the way for breeders to engineer new crop varieties.
A new platform for speeding up and controlling the evolution of proteins inside living plants has been developed by a KAUST-led team.
Previously, this type of directed evolution system was only possible in viruses, bacteria, yeast and mammalian cell lines. The Saudi research—part of KAUST’s Desert Agriculture Initiative—has now expanded the technique to rice and other food plants. It means that plant breeders now have an easy way to rapidly engineer new crop varieties capable of withstanding weeds, diseases, pests and other agricultural stresses.
“We expect that our platform will be used for crop bioengineering to improve key traits that impact yield and immunity to pathogens,” says group leader Magdy Mahfouz. “This technology should help improve plant resilience under climate change conditions.”
To experimentally build their directed evolution platform, Mahfouz and his colleagues used a combination of targeted mutagenesis and artificial selection in the rice plant, Oryza sativa. They took advantage of the gene-editing tool known as CRISPR to generate DNA breaks at more than 100 sites throughout the SF3B1 gene, which encodes a protein involved in the processing of other gene transcripts. After manipulating the DNA of small bundles of rice cells in this way, the researchers then grew the mutated seedlings in the presence of herboxidiene, a herbicide that normally targets the SF3B1 protein to inhibit plant growth and development.
This strategy ultimately yielded more than 20 new rice variants with mutations that conferred resistance to herboxidiene to varying degrees. In collaboration with Stefan Arold’s group at the KAUST Computational Bioscience Research Center, Mahfouz and his colleagues then characterized the structural basis of the resistance—showing, for example, how particular mutations helped destabilize herbicide binding to the SF3B1 protein.
Herboxidiene is not widely used in industrial agriculture, but the same basic directed evolution strategy could now be used to design crops resistant to more common weed-killers. The herbicides would then eliminate unwanted surrounding plants while leaving the desired cultivated crop intact.
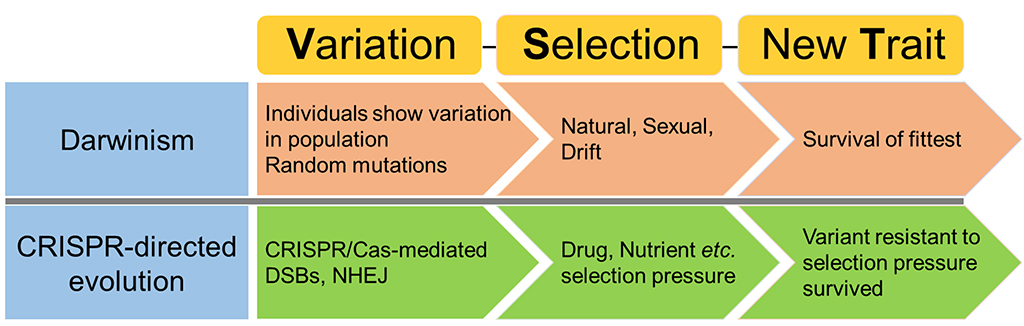
Breeders could also begin to evolve practically any trait of interest, notes Haroon Butt, a postdoctoral fellow in Mahfouz’s lab. “This is a proof-of-principle study with wide applicability,” says Butt, the first author of the paper that outlines the technology. “Our platform mimics Darwinism, and the selection pressure involved helps enforce the development of new gene variants and traits that would not be possible by any other known method.”
Read the paper: Genome Biology
Article source: KAUST – King Abdullah University of Science and Technology
Image: KAUST








