
Plants have evolved fiendishly complicated metabolic networks. For years, scientists focused on how plants make secondary metabolites, the compounds that plants produce to enhance their defense and survival mechanisms.

Plants have evolved fiendishly complicated metabolic networks. For years, scientists focused on how plants make secondary metabolites, the compounds that plants produce to enhance their defense and survival mechanisms.

Hundreds of naturally occurring specialty fatty acids (building blocks of oils) have potential for use as raw materials for making lubricants, plastics, pharmaceuticals, and more—if they could be produced at large scale by crop plants. But attempts to put genes for making these specialty building blocks into crops have had the opposite effect: Seeds from plants with genes added to make specialty fatty acids accumulated dramatically less oil. No one knew why.

Anthraquinones are a class of naturally occurring compounds prized for their medicinal properties, as well as for other applications, including ecologically friendly dyes. Despite wide interest, the mechanism by which plants produce them has remained shrouded in mystery until now.

A group of scientists from Sechenov University, Russia, and La Trobe University, Australia, have developed a fast and cost-effective method of detecting and identifying bioactive compounds in complex samples such as plant extracts. They successfully applied the method to examine Mediterranean and Australian native culinary herbs. Three articles on this work were published in Applied Sciences, Journal of Pharmaceutical and Biomedical Analysis and Journal of Chromatography A.
Since ancient times, people have been using herbs as food additives and medicines, though a search for useful compounds and a study of their properties remain a difficult task. It is possible to examine a compound if it is stable enough and can be separated from other substances in a sample. However, plant extracts contain hundreds of compounds. In the past, only known compounds were investigated by target analysis and most bioactive compounds were left undiscovered. Thus, the number of compounds that are yet to be explored is so huge that methods that can both screen mixtures and identify the compounds responsible for bioactivity are of greater value.
The authors of the papers used an Effect Directed Analysis (EDA) approach, which is a combination of chromatographic separation with in situ (bio)assays and physico-chemical characterisation to discover and identify bioactive compounds in complex plant samples. Thin-layer chromatography (TLC) and high performance thin-layer chromatography (HPTLC) are well established, chromatographic separation techniques ideally suited for high-throughput screening of bioactive compounds in crude samples.
To separate substances, TLC uses the fact that various compounds are transported by a solvent and absorbed by a sorbent at different speeds. A sorbent-coated plate with a studied mixture is immersed with one end in the solvent, and under the action of capillary forces, it begins to rise along the plate, taking the substances of the mixture with it. As they move upward, the compounds are absorbed by the sorbent and remain as horizontal bands that can be distinguished in visible, infrared or ultraviolet light. Using this method, crude extracts can be analysed directly with no preparation and possible loss of sample components.
Bioassays allow to determine the properties of compounds, such as toxicity, observing how model organisms (bacteria, plants or small animals) react to them. In this way, one can select extracts able to inhibit the action of individual enzymes or reactive oxygen species.
Combination of TLC chromatography with microbial (bacteria and yeast) tests and biochemical (enzyme) bioassays enables rapid and reliable characterization of bioactive compounds directly on the chromatographic plates, without isolation/extraction. The advantage of HPTLC is that plates/chromatograms can be directly immersed into enzyme solution (bioassays), incubated for up to several hours, followed by visualization of the (bio)activity profile via an enzyme substrate reaction as bioactivity zones. This approach is more cost effective, enabling a more streamlined method to detect and characterise natural products that are suitable candidates for further investigation as potential new drug molecules.
Using this method, scientists examined the properties of bioactive compounds from culinary herbs commonly used in the Mediterranean diet: basil, lavender, rosemary, oregano, sage and thyme. Australia’s native plants were added to the list: lemon myrtle (Backhousia citriodora), native thyme (Prostanthera incisa), sea parsley (Apium prostratum), seablite (Suaeda australis) and saltbush (Atriplex cinerea). Some of the secondary metabolites from these plants exhibit significant antioxidant activity and enzyme inhibition, like α-amylase inhibition. Therefore, these herbs may be preventive not only against cardiovascular diseases but also type 2 diabetes. The enzyme α-amylase breaks down polysaccharides, thereby increasing blood sugar levels. Recent studies suggest that hyperglycemia induces generation of reactive oxygen species, alteration of endogenous antioxidants and oxidative stress. It was found that patients with uncontrolled sugar levels in addition to diabetes also suffer from accelerated cognitive decline independent of their age. Although Australian native herbs are used as a substitute for related European plants, their medicinal properties are much less studied.
After preparing the extracts, the scientists began to study their composition and qualities. Rosemary and oregano extracts showed the greatest antioxidant activity, while sage, oregano and thyme were the best at slowing down reactions involving α-amylase (extracts from lavender flowers and leaves were the only ones not to show this effect). Among the studied Australian native herbs, lemon myrtle showed the strongest antioxidant properties, with the best α-amylase inhibition observed with extracts of native thyme (this property was noticed for the first time), sea parsley and saltbush.
The study of plant extracts using bioassay and thin-layer chromatography allows scientists to examine a variety of compounds, find mixtures that have the desired properties and isolate substances that exhibit them to the greatest extent. This fast and cost-effective method will be useful for finding new drug compounds.
Read the papers: Applied Sciences, Journal of Pharmaceutical and Biomedical Analysis and Journal of Chromatography A.
Article source: Sechenov University via Eurekalert
Image credit: Melburnian / Wikimedia

Researchers show that the wallflower is an excellent model plant for discovering new cardenolides that could be used to treat heart disease and cancer.
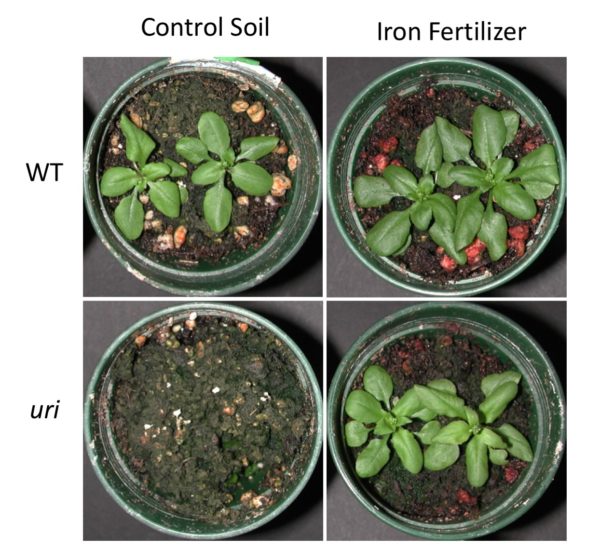
Researchers have discovered a gene that controls the regulation of iron uptake in plants, according to a new study. With over 2 billion people suffering from iron deficiency around the world, the discovery could be the key to increasing the iron potency of crops.

Vitamin E is a potential antioxidant that could act as a sentinel in plants, sending molecular signs from chloroplast –a cell organelle- to the nucleus. This flow of information reaching the cell nucleus –retrograde signalling- is a molecular mechanism that would ease the adaptive response of plants in physiological stress situation (salinity, lack of nutrients, drought, senescence, etc.).

Zostera marina. Public domain, via Wikimedia Commons.
It’s the ancient story of plant evolution: photosynthetic algae moved to damp places on land, eventually evolving more complex architecture, and spreading across almost all terrestrial habitats. To cope with the drier conditions, plants developed roots to absorb water, and vascular tissue to transport it; a waxy cuticle coating their surfaces to prevent evaporation; and microscopic pores called stomata that open to allow carbon dioxide to diffuse in for photosynthesis but close to prevent excessive water loss.
How, then, does eelgrass (Zostera marina) fit in to this tale? It’s a monocot descended from the flowering plants, but it has turned its back on dry land and returned to the sea; a rare feat that only appears to have happened on three occasions. The recent sequencing of the eelgrass genome has revealed several interesting insights into the dramatic genetic changes that have allowed it to adapt to what lead author Professor Jeanine Olsen described as, “arguably the most extreme adaptation a terrestrial (and even a freshwater) species can undergo.”
Sayonara to stomata
If you live in the sea, conserving water isn’t your main concern. Eelgrass was known to lack stomata, but genetic comparisons to other species, including its freshwater relative Spirodela polyrhiza, revealed the first surprise of the study: eelgrass has lost not only its stomata but also the genes involved in their development and patterning. “The genes have just gone, so there’s no way back to land for seagrass,” said Olsen.
A difference in defense
When angiosperms are attacked by herbivores or pathogens, their defense response typically involves the release of volatile secondary metabolites through their stomata. How can eelgrass release these compounds without stomata? The answer is: it doesn’t. The genome study found that eelgrass is missing crucial genes involved in making ethylene (an important hormone release in times of stress), as well as those responsible for producing non-metabolic terpenoids, which act to repel pests.
Selective pressures of the marine environment differ greatly from those of terrestrial habitats, so different pathways may be involved. Second, eelgrass has a wide repertoire of pathogen resistance genes, which suggests that it is exposed to a very different set of pathogens that may not respond to typical immune responses. Third, volatile secondary metabolites are often involved in attracting pollinators; this is not believed to be necessary in eelgrass, where submarine pollination occurs using the water itself.

Zostera marina – National Museum of Nature and Science, Tokyo. Public domain, CC0 1.0, via WikiMedia Commons.
Changing the cell wall
Eelgrass is subject to extremely salty conditions, and it’s had to adapt to osmotic stress. Unlike typical plant cell walls, eelgrass has engineered its cell wall matrix to retain water in the cell wall, even during low tide. This involves depositing sulfated polysaccharides and low methylated pectins in the cell wall matrix, but until its genome was sequenced no-one knew exactly how. It turns out that eelgrass has rearranged its metabolic pathways: “They have re-engineered themselves,” Olsen explains.
Living with a lack of light
Some species of Zostera can grow in water 50m deep, where light levels are reduced and shifted into a narrow wavelength range; ultraviolet (UV), red and far-red light have particularly low penetration after the first 1–2m of seawater. In a classic eelgrass ‘use it or lose it’ response, it has lost the UVR8 gene, which is responsible for sensing and responding to UV damage, as well as the phytochromes associated with red and far-red receptors. It does, however, retain the photosynthetic machinery, including photosystems I and II.
Unravelling angiosperm evolution
The recent eelgrass publication has revealed how this plant has either lost or adapted typical angiosperm traits to suit its needs, by ditching its stomata, volatile secondary metabolites and certain light sensing genes, or by altering the structure and function of the cell wall. It also developed adaptations that enable gas exchange, help pollen stick to submerged stigmas, and promote nutrient uptake.
Could these adaptations be useful in crop breeding? While a lack of defense compounds would probably be a step backwards, it would be extremely useful to understand how eelgrass copes with biotic stresses without them. Removing light receptors would also be problematic, but could eelgrass help us to develop crops that can grow in shaded conditions, perhaps in intercropping systems? What can we learn from eelgrass’ nutrient uptake and salt-tolerant adaptations?
Now that we have seen some of the secrets of eelgrass, how can we best make use of them?
Read the paper: The genome of the seagrass Zostera marina reveals angiosperm adaptation to the sea (Open Access)
Read the editorial: Genomics: From sea to sea (paywall)
Read the press release: Genome of the flowering plant that returned to the sea