Biologist discovered that repeats in genetic patterns associated with plant immunity may cause its immune system to misbehave.
Genomic information from plants can be used to enhance agricultural production and improve food security in a sustainable manner. Through better understanding of the relationship between a plant’s genetic information and the resulting behaviour, improved crops with better traits (resistance to diseases, tolerance to drought, etc.) can be developed. This can be achieved through cross pollinating plants with selected traits to produce a hybrid with the desired characteristics.
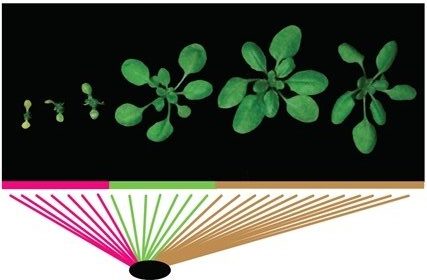
A research team led by Prof Eunyoung CHAE from the Department of Biological Sciences, NUS has discovered that in selective plant breeding, the genes responsible for providing defence responses against powdery mildew (RPW8) in the Arabidopsis thaliana plant, when duplicated as multiple copies in the genome, can misregulate immune receptors to trigger autoimmunity in hybrid plants. This can cause the premature gradual death of the foliage in these hybrids, known as hybrid necrosis.
The researchers applied a technique known as the genome-wide association mapping study (GWAS) to the model plant Arabidopsis thaliana in this research. GWAS is an important tool to identify genetic variation in plants which are associated with a particular trait. It allows the researchers to identify Single Nucleotide Polymorphisms (SNPs) which mark the causal natural variation in the genome responsible for growth-immunity trade-off. These SNPs are like postal codes showing the genetic information that are associated with autoimmune symptoms in plants. The SNPs were found to be located adjacent to a multi-gene cluster which is known to confer resistance to powdery mildew (a fungal disease that affects a wide range of plants). The team found that having multiple copies of this set of genetic codes (RPW8) in Arabidopsis thaliana is detrimental as it triggers its autoimmunity mechanism even in the absence of pathogens.
Prof Chae said, “The plant Arabidopsis thaliana used in our study belongs to the Brassicaceae family, and the family members include vegetables such as broccoli, kale, cabbage, mustard and bok choy. Our team is currently assessing the variability of the plant immune system, particularly on the powdery mildew resistance gene clusters in this family, and if the variability is associated with heightened resistance and/or trade-off between immunity and growth.”
Read the paper: PLOS GENETICS
Article source: National University of Singapore
Image credit: PLOS GENETICS













