


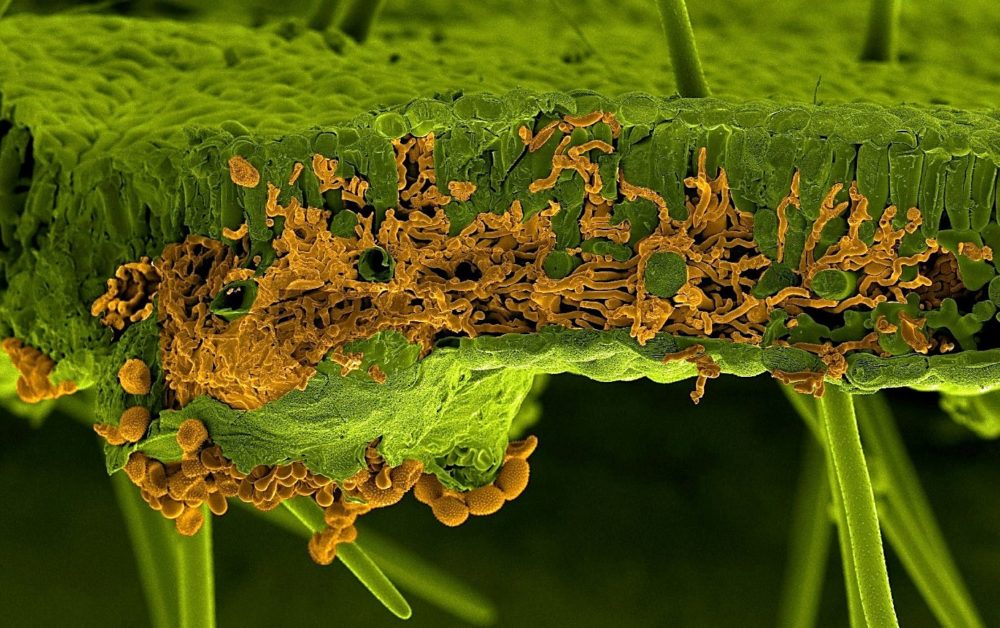
An international research collaboration has successfully assembled the complete genome sequence of the pathogen that causes the devastating disease Asian soybean rust.
The research development marks a critical step in addressing the threat of the genetically-complex and highly-adaptive fungus Phakopsora pachyrhizi which has one of the largest genomes of all plant pathogens. Asian soybean rust has a devastating impact on soybean, an internationally important crop with 346 million tonnes produced globally.
In conditions favourable to its spread, the rust can destroy up to 90% of the soybean harvest. At present soybean growers in major areas of cultivation such as Latin America must use chemicals to protect crops.
The largest producer of the soybean is Brazil, where the combined cost of losses and disease control measures is US $2 billion per season.
The new dataset comprises the genome sequence of three isolates (K8108, MG2006 & PPUFV02) of which one has been assembled at chromosome level detail (PPUFV02). These three genomes will be hosted by the Joint Genome Institute and will be made available soon.
The three genomes will be repeat masked and annotated in the same way, to facilitate direct comparisons and inferences for the research community. This will enable researchers to study the molecular mechanisms of the pathogen, paving the way for breeding and engineering of disease-resistant crops.
The international consortium behind the project comprised 11 research and industry partners: The 2Blades Foundation, KeyGene, the Joint Genome Institute (JGI), Bayer, Syngenta, the Brazilian Company of Agricultural Research (Embrapa), l’Institut National de la Recherche Agronomique (INRA – France), the German Universities of Hohenheim and RWTH Aachen, The Sainsbury Laboratory, and the Federal University of Viçosa (Brazil).
The soybean rust pathogen is highly adaptive and disease resistance genes present in soybean have been overcome rapidly, and the pathogen is building resilience against the current generation of fungicides. These two factors leave few solutions for controlling the disease in the field.
Two of the three isolates that the consortium have sequenced are from Brazil, where the impact of soybean rust is a huge problem for farmers.
Phakopsora pachyrhizi has a highly complex genome, it is 60 times bigger than the yeast genome, composed of 93% repetitive elements and possesses two nuclei.
This complexity has delayed progress on the sequencing of this pathogen, and meant that high-end, next‐generation sequencing technologies were required to complete the task.
Given the importance of this disease, KeyGene made their PromethION machine (an industry first) and their sequencing and bioinformatics experts available pro bono. This way ultra-long DNA-sequencing reads of the pathogen and a high quality nanopore assembly were produced. This allowed Dr. Yogesh Kumar Gupta from the 2Blades group to generate a chromosome level assembly of the isolate PPUFV02, of which the DNA was provided by their long-term collaborator Prof. Sérgio Brommonschenkel at the Federal University of Viçosa (Brazil).
The consortium has also generated a transcriptome atlas of all the fungal structures and infection stages of the pathogen.
“Asian soybean rust is a critical challenge for soybean growers,” said Dr. Peter van Esse, leader of the 2Blades Group at The Sainsbury Laboratory, Norwich, one of the collaborators.
“A chromosome level genome assembly allows the scientific community to study, in unprecedented resolution, components of the pathogen that are critical for causing disease. This is a critical first step towards the design of transformative control strategies to combat this highly damaging pathogen.”
Peter van Esse
Access the sequence: Mycocosm
Article source: The Sainsbury Laboratory
Image credit: U. Steffens, Bayer Crop Science
This week’s post was written by Jonathan Ingram, Senior Commissioning Editor / Science Writer for the Journal of Experimental Botany. Jonathan moved from lab research into publishing and communications with the launch of Trends in Plant Science in 1995, then going on to New Phytologist and, in the third sector, Age UK and Mind.
In this week of the XIXth International Botanical Congress (IBC) in Shenzhen, it seems appropriate to highlight outstanding research from labs in China. More than a third of the current issue of Journal of Experimental Botany is devoted to papers from labs across this powerhouse of early 21st century plant science.
Collaborations are key, and this was a theme that came up time again at the congress. The work by Yongzhe Gu et al. is a fine example, involving scientists at four institutions studying a WRKY gene in wild and cultivated soybean: in Beijing, the State Key Laboratory of Systematic and Evolutionary Botany at the Institute of Botany in the Chinese Academy of Sciences, and the University of the Chinese Academy of Sciences; and in Harbin (Heilongjiang), the Crop Tillage and Cultivation Institute at Heilongjiang Academy of Agricultural Sciences, and the College of Agriculture at Northeast Agricultural University. Interest here centers on the changes which led to the increased seed size in cultivated soybean with possible practical application in cultivation and genetic improvement of such a vital crop.
Botanic gardens are also part of the picture. In another paper in the same issue, Yang Li et al. from the Key Laboratory of Tropical Plant Resources and Sustainable Use at Xishuangbanna Tropical Botanical Garden in Kunming (Yunnan) and the University of the Chinese Academy of Sciences in Beijing present research on DELLA-interacting proteins in Arabidopsis. Here the authors show that bHLH48 and bHLH60 are transcription factors involved in GA-mediated control of flowering under long-day conditions.
Naturally, research on rice is important. Wei Jiang et al. from the National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University (Wuhan) describe their research on WOX11 and the control of crown root development in the nation’s grain of choice, which will be important for breeders looking to increase crop yields and resilience.
The other work featured is either in Arabidopsis or plants of economic importance: Fangfang Zheng et al. (Qingdao Agricultural University, also with collaborators in Maryland) and Xiuli Han et al. (Beijing); Yun-Song Lai et al. (Beijing/Chengdu – cucumber), Wenkong Yao et al. (Yangling, Shaanxi – Chinese grapevine, Vitis pseudoreticulata), and Xiao-Juan Liu et al. (Tai-an, Shandong – apple).
Shenzehn has grown rapidly and is now highly significant for life science as home to the China National GeneBank (CNGB) project led by BGI Genomics. The vision as set out by Huan-Ming Yang, chairman of BGI-Shenzhen, is profound – from sequencing what’s already here, often in numbers per species, to innovative synthetic biology.
Shenzehn is also home to another significant institution, the beautiful and scientifically important Fairy Lake Botanic Garden. At the IBC, the importance of biodiversity conservation for effective, economically focused plant science, but also for so many other reasons to do with our intimate relationship with plants and continued co-existence on the planet, was a central theme.
The research highlighted in Journal of Experimental Botany is part of the wider, positive growth of plant science (and, indeed, botany) not just in China, but worldwide. The Shenzehn Declaration on Plant Sciences with its seven priorities for strategic action, launched at the congress, will be a guide for the right development in coming years.